This is something I've been chewing on for more than a year now and yet never got myself to blog about (although I have mentioned in private or in comments here and there). Impelled by the minor but quite apparent NE African influence, genetic and cultural, on the Neolithic peoples of the Levant, whose offshoots eventually landed in Greece triggering the European Neolithic, I decided in the Spring of 2014 to explore, via mass-lexical comparison, if Basque language (and by extension the wider Vasconic family, which I believe now to be that of mainline European Neolithic) might have any relation with Nubian languages. I did not expect to find anything but noise but to my surprise the number of apparent cognates is quite significant.
My primary analysis was
this one but now I have combined it with a comparison with Proto-Indoeuropean (PIE), which is also very probably related to the roots of Vasconic:
LINK (open office spreadsheet).
The synthesis is as follows:
Of course the "cognates" are only apparent cognates at this stage of the research and the evaluation is necessarily subjective. But judge yourselves.
If we discard the "weak" apparent cognates, the vocabulary correlation between Basque and Nubian and between Basque and PIE is pretty similar. But, in my understanding, both are well above the noise threshold, an example of which could be the PIE-Nubian apparent cognates, which are many many less.
I must say anyhow that the oblique apparent cognates, that is when one word sounds much not like its strict synonym but a related one (for example words meaning hot and fire), look all very solid and most intriguing.
Also, when attributing probabilities to origins of Basque words, Nubian appears to be at the origin of almost double the words (26%) that can be attributed to PIE (15%). Of course, for lack of data or because they actually have other origins, the unknown origins apply to the majority of words (56%), double than the Nubian origin ones.
However Nubian here is constituted of three different languages (Dilling, Nobiin and Midob), while PIE is just a single theoretical construct. This last must be done this way because many modern and historical IE languages, notably in Europe, have other Vasconic substrate influences, which must be studied separately from general PIE-Vasconic shared vocabulary. This kind of late Vasconic influence is very much unlikely in the case of Nubian instead. In any case I don't know of any a proto-Nubian Swadesh list readily available.
Finally I must mention that because the PDF format is horrible for copy-pasting, I chose to re-transcribe the Nubian words according to my best approximation using a normal keyboard (not always the same characters that the original list uses).
Strongest Basque-Nubian apparent simple cognates
- Basque - Nubian languages (English)
- azal - àzì, àzzì-di (bark)
- haragi - árízh (meat)
- odol - ógór, èggér (blood)
- buru - úr (head)
- oin - ó:y (foot)
- esku - ish-i, ès-sì (hand)
- hil* - di-ìl (to die)
- euri - are, ara, áwwí, áré, árí, áró (rain)
- harri - kugor, kakar (stone) [notice also the pre-IE root *kharr- speculated to be at the origin of Karst, etc.]
- lur - gùr (soil, ground)
- haize - irsh-i, éss-í (wind)
There are some others that are shared with Indo-European and with similar subjective "weight", not listing them here to keep things clear. There are also other apparent cognates that are arguably less clear like bat - be (one) that I'm also skipping here but you can find in the spreadsheet.
*Hil (meaning both to die and to kill in Basque, which can't be confused because they conjugate differently) seems ancestral to English ill and kill (this one via a Germanic precursor).
The intriguing oblique cognates
Notice that these words do not mean the same, yet their meanings seem strikingly related.
- Nubian (English) - Basque (English)
- hor, koy, kà:r (tree) - harri (stone) [notice that zuhaitz (tree) can be interpreted etymologically as zur-haitz = wood-rock, so the relation is not that weird]
- ok-i, og (breast) - ogi (bread)
- a-l (heart) - ahal (can (verb), potential, power)
- azh, àz-ír, àzza (to bite) - (h)ortz (tooth), aitz (rock, peak) [some argue that originally "to cut", present in many cutting tool names: aizkor = axe, aitzur = hoe, aizto = knife, etc.*]
- shu, zhúù (to walk) - joan (to go) [often pronounced jun or shun]
- é:zhi (water) - heze (wet) [also archaic particle *iz-, meaning "water" by all accounts: itxaso = sea, izurde = dolphin, izotz = ice, and common in Vasconic river toponymy]
- zhuge (to burn) - su (fire)
- zhùg, sù, sú:w (hot) - su (fire)
- úr-i, úrúm (black) - urdin (blue) [archaic also green, grey]
*This one is an obvious and very prevalent Vasconic substrate infiltrator in Western IE languages: axe, adze, azada (hoe in Spanish), etc.
How can this be possible?
It is of course a mere working hypothesis and ultimately you judge but I find it hard to disdain. However there is no apparent connection, notably no significant genetic connection, between Basques and Nubians. So how can we explain this?
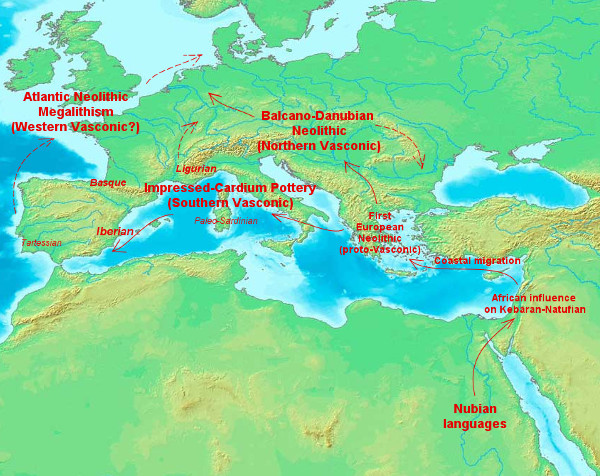
I have it reasonably clear myself, so I made a map to explain it:
Basque is after all just the last survivor of a once much larger family (Vasconic), a family that most likely corresponds to the languages spoken by the early European farmers (mainline Neolithic of Aegean roots). As that expansion was largely done in about a mere thousand years, I estimate that when both branches met near the Rhine, the two peoples could still understand each other, even if with some difficulty. Only the Southern/Western branch(-es) survived long enough to leave historical evidence, so it is hard to guess how the Northern branch evolved anyhow.
The Nubian linguistic connection is anyhow not the only thing that requires the Levant or Palestinian Neolithic step, also Y-DNA E1b-M78 (mostly V13 in Europe, attested in some early farmers and still very important among Greeks and Albanians particularly) and probably the so-called "Basal Eurasian" component that Lazaridis detected among early European farmers and that could well be the signature of African genetics from the Nile.
Linguistically, also the very notorious presence of Semitic (an Afroasiatic branch) in West Asia is surely another legacy of the same African influences in the Mesolithic Levant. Before this research, I thought it was the only one but now I strongly suspect that at some point Nubian (Nilo-Saharan) languages were also present in the region. Maybe one (Nubian evolving towards Vasconic) corresponded to Natufian proper and the other (proto-Semitic) to Harifian, the semi-desert pastoralist facies of the same wider culture. Can't say for sure.
The chain was once long but now only some of the most distant links remain unbroken. It is difficult to imagine that they were ever connected at all...
To do...
A lot remains to be done, of course:
- These mass lexical comparisons only apply to a few families in the region and the rest should also be tested for. My energies are limited and so are my qualifications as "linguist", so I encourage others, hopefully more energetic and knowledgeable, to expand.
- Grammatical features cannot be analyzed by this methodology. Again my means are limited.
- Anthropological research would be an interesting complement. So far the only shared cultural trait I could spot would be the use of bells attached to ankles for dancing but there could be others.
- ...