This is the second attempt at discussing a very interesting paper which has been hurt by an editor error in publication (a key informative element, table 2, has its columns all swapped).
I realized that something looked quite wrong and notified the authors, who are now awaiting for PLoS to correct the problem. In the meantime they have been so kind as to provide me with a copy of the original PDF manuscript so I could properly collate the haplogroup data and share it with readers of this blog.
Ganesh Prasad Arun Kumar et al., Population Differentiation of Southern Indian Male Lineages Correlates with Agricultural Expansions Predating the Caste System. PLoS ONE 2012. Open access → LINK [doi:10.1371/journal.pone.0050269]
The authors took special interest into sampling tribes, some of which
are still foragers and a reference for all kind of anthropological
research of South Asia, all Eurasia and even beyond. They also sorted
the various populations into groups or classes based on socio-economic
reality (and language in some cases) rather than the, arguably
overrated, varna (caste) system.
The categories used are:
- HTF - Hill Tribe Forager (foragers of Tamil or Malayalam language)
- HTK - Hill Tribe Kannada (foragers of Kannada language)
- HTC - Hill Tribe Cremation (tribals who cremate their dead, not sure if silviculturalists)
- SC - Scheduled Castes (castes traditionally discriminated against, Dalits)
- DLF - Dry Land Farmers
- AW - Artisan and Warrior related castes
- BRH - Brahmin-related castes with irrigation farming economy
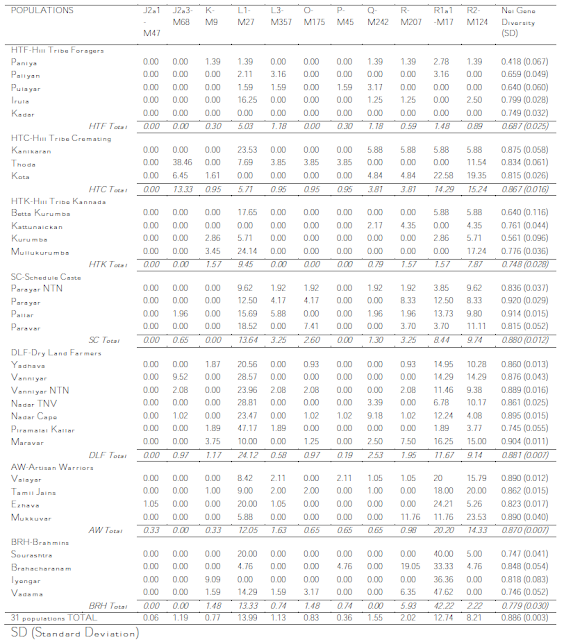
And, as I said then, the bulk of the data is in table 2, which I have the privilege of sharing with you as it really is (in two blocs, as it was in the PDF):
And now finally I can get to discuss the details with the certainty of talking about real data.
Haplogroup C
As the authors note, 90% (66/74) of all the C-M130 samples belong to C5 (M356), while the rest (8/74) tested negative for both C5 and C3 (M217), so I guess we are here before at leas one other subhaplogroup of C (because the likelihood of being Japanese C1 or Australasian C2 or C4 is practically zero).
The eight C* individuals are scattered (table S1) among several groups (all of which also display C5, as well as F*) but notably concentrated among the Piramalai
Kallar (4/5 within C),
which are a DLF group (corrected upon comment).
Besides C*, which may well be a remnant of either the early Eurasian expansion or of the first backflows from SE Asia (a likely not-so-likely candidate for the origin of macro-haplogroup C), the very notable presence of C5 among tribals and some farmers may well indicate that the origin of C5 is in South Asia, even if the clade also has some presence in Central and West Asia.
Haplogroup C has a high variance in this study (0.80), greatest among DLF (0.89) and HTF (0.81).
(Update: see also appendix below).
Haplogroup E
As we should expect, this lineage of African origin (with important presence in West Eurasia) is only found at low levels among farmers (DLF). It may well be a remnant of early Neolithic flows, being strongly linked with Neolithic in the case of Europe for example.
Paragroup F*
The most striking thing about Paragroup F*, i.e. F(xG,H,J,K), is that it is found at such high numbers and very especially so among the hunter-gatherers, where it is often the main lineage (or lineages). It is also important among dry land farmers and the Valayar (AW class) but it is rare to non-existent among the other caste groups, which may represent relatively recent arrivals.
Something that this confirms, along with other older data about F basal diversity, is that the main Eurasian Y-DNA haplogroup, which is of course F itself, coalesced necessarily in South Asia.
Said that, I cannot underline enough how relevant is to find rare F sublineages (i.e. F* - so rare that have not even been properly identified by downstream markers yet) among the last forager peoples of South Asia, often as dominant clade.
Haplotype neighbor-joining exercise was performed however, indicating founder effects (possible new haplogroups to be yet described) among tribals:
However it is also obvious that there is a lot of diversity as well. In fact, paragroup F* does have a high variance in Tamil Nadu (0.81), being highest again among the DLF class (0.85).
Haplogroup G
Haplogroup G does exist in South Asia and this paper makes it evident. More so, its distribution in Tamil Nadu includes some foragers and other tribals, although it is more common among "Neolithic" classes.
Among these the Ivayengar (BRH) show almost 27% (3/11), however other BRH populations do not show any G, while the DLF ones instead all have relevant G. Therefore this lineage may tentatively be associated in Tamil Nadu with the Neolithic.
Haplogroup G, suggested by the authors to be a Neolithic arrival, has an strikingly high variance in Tamil Nadu (0.83) with top level among the following classes: AW (1.05), SC (0.94) and BRH (0.82).
Even if the distribution corresponds well with a Neolithic inflow the diversity is surprisingly high and it tells me that more research is needed about this lineage in South Asia. After all it is one of the basal descendants of F, whose coalescence took place no doubt in the subcontinent.
Haplogroup H
Haplgroup H is of course very common in Tamil Nadu but it must be noticed that it is concentrated in the H1(xH1a) category, as well as some notable H(xH1,H2), which tends to weight in favor of a southern ultimate origin of this important South Asian clade (as also proposed in
the recent study on the Roma People).
H* is distributed among many populations, the only class fully excluded being the BHR one, which is generally considered to be a recent historical arrival from the North (mostly confirmed by genetics). Some tribes have the highest values but then some others totally lack it.
H* has extremely high variance levels in Tamil Nadu (1.33), being highest among the SC class (1.46), followed by the AW one (1.18) and the DLF one (0.91). This is totally consistent with a South Asian origin of H overall.
H1* is standard issue in all populations. The highest values are among the Kannada-speaking tribals (HTK), followed by cremation-practicing tribals (HTC).
H1a instead is only found in one population at very low levels, strongly suggesting that this clade is not from the region. H2 instead is found at low levels among many groups.
Unlike H*, H1 and H2 have rather low diversity levels in Tamil Nadu: 0.41 and 0.59 respectively.
Haplogroup J
J(xJ2) is found at anecdotal levels in a couple of lower class populations (one tribal and the other SC). It would be particularly interesting if we knew it is not J1 as well - but we don't.
Most is J2(xJ2a) although J2a3 is also important among several populations.
It is generally believed that J in South Asia is of Neolithic origin and I will not question it but still... notice how important it is among several tribal foragers: >4% in four tribes, levels on average similar to those of farmers and Brahmins.
J2* is rather high in diversity (0.73), notably among the AW class (1.0), while J2a3 is very low instead (0.29).
Paragroup K(xL,R)
Or if you wish paragroups K* and P*, as well as haplogroups O and Q.
The always interesting K* is found at low levels among some tribals and most DLF populations. However the peak is among Viyengar Brahmins. May it be haplogroup T?, L2?
O in this area is almost for sure O2a brought by Austroasiatic-speaking rice farming tribes in the Neolithic. It is found at low levels in some groups, including the Thoda "cremation tribals" (who look quite "Neolithic" also because of their high levels of J2).
P(xQ,R), which is most common towards Bengal, is found in Tamil Nadu at low levels among diverse populations. On first impression I'd say it's also a Neolithic influence although, of course, in the wider subcontinental region it must be much much older.
Q is found at low levels in diverse populations being maybe somewhat more common among the Scheduled Castes class.
Haplogroup L
Haplogroup L is an important South Asian lineage with penetration in West and Central Asia and a center of gravity around Sindh (Pakistan), although it is also very common in West and South India.
In Tamil Nadu L1a (L1 in the table) is common among nearly all sampled populations with peak among the dry land farmers' class.
Instead L1c (formerly L3) is relatively rare, peaking among the Scheduled Castes class. No mention is made of any other L.
Both clades show low variance in the region (0.41 and 0.22 respectively), consistent with their origin being further North.
Haplogroup R
R(xR1a1,R2) is found in several populations at non-negligible levels: near 5% among some tribals, 8% among the Parayar (SC) and the Maravar (DLF), also 12% Mukkuvar (AW) and as much as 19% among some Brahmins (the Brahacharanam who are also high in P*). This could well be R*, R1*, R1a*, R1b, etc. and indicates in my understanding target populations for future research on the hot topic of the ultimate origins of R1 and R1a (see also
here).
R* shows clearly high variance: 0.97 on average, being highest among the DLF class (1.25), followed by the BRH class (0.99)
R1a1, which may well be related to Indoeuropean expansion (or just Neolithic or whatever, better resolution is needed especially in Asia) is found at very high levels among the BRH class (45%), followed by the AW one (20%) other classes show near 10% except the hunter-gatherers (HTF and HTK) who have only anecdotal presence of this lineage.
R1a1 shows rather low variance (0.41), rather confirming its immigrant origin from North India (incl. maybe Pakistan, Bangla Desh, Nepal...). All classes are similar for this value.
R2 (a South Asian lineage with occasional offshoots into West and Central Asia) is common in all groups except the HTF class. The highest levels (c. 15% avg.) are among cremating tribals and artisan/warrior classes. I'd say that with the likely origin of R2 somewhat to the North of this region, it seems normal that
Kannada-speaking tribals (HTK, who must be immigrants from Karnataka or at least strongly influenced by this other Dravidian country's culture) have lots of it, while the more locally native HTF almost lack it instead.
R2 shows mid-level diversity on average (0.65) but the HTK class displays very high diversity for this lineage (1.05).
Different interpretations
Notice that my take and that of the authors on the autochthonous nature of each of the lineages may vary or be debatable. They say the following:
The geographical origins of many of these HGs are still debated.
However, the associated high frequencies and haplotype variances of HGs
H-M69, F*-M89, R1a1-M17, L1-M27, R2-M124 and C5-M356 within India, have
been interpreted as evidence of an autochthonous origins of these
lineages during late Pleistocene (10–30 Kya), while the lower frequency
within the subcontinent of J2-M172, E-M96, G-M201 and L3-M357 are viewed
as reflecting probable gene flow introduced from West Eurasian Holocene
migrations in the last 10 Kya [6], [7], [16], [23].
Assuming these geographical origins of the HGs to be the most likely
ones, the putatively autochthonous lineages accounted for 81.4±0.95% of
the total genetic composition of TN populations in the present study.
Mostly our differences stem on my doubts about the real origins of R1a1 (which could well be West Asian by origin) and that I imagined L1c (aka L3) as native from South Asia (uncertain now admittedly). But otherwise I agree. The hottest issue is no doubt the origin of R1a or R1a1, still unsolved.
PC Analysis
A quick visual understanding of the relations between the different classes can be obtained from figure 2:
Check table 1 for population codes but essentially: squares are tribes and circles caste populations; red are the HTF class, green the HTK and yellow the Brahmin-related groups (BRH).
These are the outliers: all the rest, including HTC, cluster together near the (0,0) coordinates.
It is also clearly indicated in Fig. 2A how R1a1, H1 and F* are the strongest defining markers.
Old structure
As always, take age estimates, also provided, with utmost caution and distrust. However I must mention that the main conclusion of the authors is that the haplogroup structure in the region pre-dates the introduction of the caste system as such and is, in their opinion, of Neolithic age.
__________________________ . __________________________
Appendix (update Dec 2):
Much of the discussion below has been
on the origins of haplogroup C. I have been pointed to
Hammer 2006 and this haplotype NJ tree (fig 4d) of what was known back in the day as C* and C1. At that time neither Australian C4 nor Asian C5 had been described yet. However Wallacean/Melanesian/Polynesian C2 and NE Asian and Native American C3 are not shown here.
Annotations (
C1,
C4 and
root?) by me:
Maybe even more interesting is Fig. 3 from
Redd 2002, which shows the whole C haplogroup tree and clearly annotates the likely root (branch to haplogroup B):
While C4 is not obvious here, the fact that South Asian (Indian subcontinent) C* is central to all the haplogroup is again underlined.
The protuberance to the top might be C5, while the one to the bottom may well correspond with the SE Asian cluster above, at least partly. The differences underline the limitations of this STR-based method alone to infer real phylogenies - but it is anyhow much better than nothing.