I will today stop at a new genetic paper that does not deal with humans but with a not-so-distant relative: house mice. Specifically with mice in the remote Kerguelen islands, which have been known to humans (and hence to mice) only since 1772.
Emilie A. Hardouin et al., House mouse colonization patterns on the sub-Antarctic Kerguelen Archipelago suggest singular primary invasions and resilience against re-invasion. BMC Evolutionary Biology 2010. Open Access.
Abstract (provisional)
Background
Starting from Western Europe, the house mouse (Mus musculus domesticus) has spread across the globe in historic times. However, most oceanic islands were colonized by mice only within the past 300 years. This makes them an excellent model for studying the evolutionary processes during early stages of new colonization. We have focused here on the Kerguelen Archipelago, located within the sub-Antarctic area and compare the patterns with samples from other Southern Ocean islands.
Results
We have typed 18 autosomal and six Y-chromosomal microsatellite loci and obtained mitochondrial D-loop sequences for a total of 534 samples, mainly from the Kerguelen Archipelago, but also from the Falkland Islands, Marion Island, Amsterdam Island, Antipodes Island, Macquarie Island, Auckland Islands and one sample from South Georgia. We find that most of the mice on the Kerguelen Archipelago have the same mitochondrial haplotype and all share the same major Y-chromosomal haplotype. Two small islands (Cochons Island and Cimetiere Island) within the archipelago show a different mitochondrial haplotype, are genetically distinct for autosomal loci, but share the major Y-chromosomal haplotype. In the mitochondrial D-loop sequences, we find several single step mutational derivatives of one of the major mitochondrial haplotypes, suggesting an unusually high mutation rate, or the occurrence of selective sweeps in mitochondria.
Conclusions
Although there was heavy ship traffic for over a hundred years to the Kerguelen Archipelago, it appears that the mice that have arrived first have colonized the main island (Grande Terre) and most of the associated small islands.The second invasion that we see in our data has occurred on islands which are detached from Grande Terre and were likely to have had no resident mice prior to their arrival. The genetic data suggest that the mice of both primary invasions originated from related source populations. Our data suggest that an area colonized by mice is refractory to further introgression, possibly due to fast adaptations of the resident mice to local conditions.
The abstract alone is quite explanatory.
I must say that the reason for the refraction may not be adaptations to local conditions as much as mere demographic pressure: any new mice would be automatic minority and have poor chances of perpetuation, even in the absence of adaptations. Numbers alone make the difference, proving statistically difficult for any new arrival to leave a mark after the first population is consolidated.
Another detail worth discussing is:
In the mitochondrial D-loop sequences, we find several single step mutational derivatives of one of the major mitochondrial haplotypes, suggesting an unusually high mutation rate, or the occurrence of selective sweeps in mitochondria.
The mutation rate does not look too striking to me: 200 years for mice is like 10,000 for us maybe, as they reach reproductive maturity in a matter of months. So it's like one (or two in a few sub-lineages) surviving mutations downstream of the founder haplotype, which is anyhow still massively dominant.
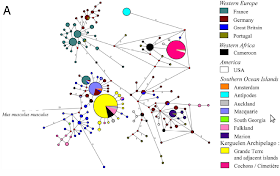 |
| Fig.2A mtDNA HVS haplotype network |
In other words: there were some distinct founder effects of some derived haplotypes, mostly in Kerguelen itself but also overseas: in Falkland and Auckland islands and even back to Europe (Britain).
The diversity of derived basal lineages clearly indicates that this Kerguelen haplotype (yellow) exploded in the islands, regardless that it may be original from mainland Africa (black) and regardless that it has also spread overseas. This in the equivalent of 10,000 human years (very roughly).
It reminds me somewhat of what we can see in Eurasian human mtDNA, with the yellow lineage resembling human mtDNA M somewhat and the magenta one like N. And that is one reason I wanted to comment this paper, really. Of course the stage of diversification and other aspects are clearly different but it does illustrates how founder effects proceed and how demic pressure alone tends to fend off new colonizations.
"It reminds me somewhat of what we can see in Eurasian human mtDNA, with the yellow lineage resembling human mtDNA M somewhat and the magenta one like N".
ReplyDeleteExactly. And note: separate regions.
"So it's like one (or two in a few sub-lineages) surviving mutations downstream of the founder haplotype, which is anyhow still massively dominant".
So doesn't that suggest that mtDNA M and N also coalesced in separate regions? The two may have left Africa together, but they must have soon parted company and not moved through South Asia together.
"So doesn't that suggest that mtDNA M and N also coalesced in separate regions?"
ReplyDeleteYes. I have sustained for long that M coalesced in South Asia and N in SE Asia. The shortest line between SE Asia and the Horn or the Nile is via South Asia (it is also the only one with a warm climate).
Anyhow, M and N must have arrived to SEA roughly about the same time, because N did not impede the advance of M in SEA and surroundings (NEA, Oceania).
It is also interesting that both mice populations have the same Y-DNA lineage anyhow.
"Anyhow, M and N must have arrived to SEA roughly about the same time, because N did not impede the advance of M in SEA and surroundings"
ReplyDeleteBut M sure impeded N's survival in South Asia.
I have to confess I have consistently carried one preconception. The conception that humans would not be too different from all other species.
I'd like to draw your attention to something about your stars that you don't seem to have noticed for yourself. In all cases, but one, all the members of the star can be arranged in geographically connected regions. Haplogroup A through northern and eastern Asia. B from the Philippines and Tengarra through most of east and SE Asia. P through New Guinea. R from New Guinea through South Asia to Europe. M through Southern Eurasia all the way from SW Asia, through South Asia and on to Australia, New Guinea and East Asia.
Similarly with all the others. Except there's a huge gap between eastern and western Ns. Unless ...
"But M sure impeded N's survival in South Asia".
ReplyDeleteNot enough.
It's not just on/off: in the early period there may have been a number of lineages, you know. Some survived and became so dominant that could not be moved, some went extinct and some managed to exile themselves into a more promising niche before suffering the same destiny.
"Except there's a huge gap between eastern and western Ns. Unless" ...
Precisely.
"Precisely".
ReplyDeleteSo how do you explain it?
Unless there's no such huge gap because all Western N sublineages (except X) are also found in South Asia (India and such).
ReplyDeleteWhen a lineage is removed geographically, it's probably because it traveled all that as a minor lineage.
You have exactly the same problem with L3 and that's where Arabia and South Asia have to be integrated: first because they are in the middle of the only logical route and second because you see archaeological and genetic evidence in those regions as well.
That's it. I think you're wasting your time and mine, sincerely.
"Unless there's no such huge gap because all Western N sublineages (except X) are also found in South Asia (India and such)".
ReplyDeletePossibly so. But in downstream mutations only, so they've come in, not originated there.
"When a lineage is removed geographically, it's probably because it traveled all that as a minor lineage".
If it travels as a minor lineage surely it's unlikely to survive.
"You have exactly the same problem with L3"
Not really. You have N haplogroups N1'5 and N2/W, and M haplogroup M1, in SW Asia. So yes, that is right on the only logical route, and you see archaeological and genetic evidence there as well.
"But in downstream mutations only, so they've come in, not originated there".
ReplyDeleteThat's a pseudo-reasoning. The same reasoning applies for the SA and WEA branches, and there are no others, so it's undetermined. There's no root (underived) haplogroups anymore, we are always looking at derived branches.
"If it travels as a minor lineage surely it's unlikely to survive".
Unlikely is not the same as impossible. There were surely many minor lineages which did not survive but some did indeed.
"Not really. You have N haplogroups N1'5 and N2/W, and M haplogroup M1, in SW Asia"...
And X, don't forget X, which is the only so phylogenetically high WEA-only lineage comparable with M1. N1'5 and N2 are shared with SA.
But it doesn't matter. Because what we do (what I do, what most geneticists do, what most other amateurs do, and they do correctly) is to consider ALL basal sublineages and not some arbitrarily chosen.
You do differently but that's the core failure in your "method" (or lack of it thereof).
You cannot understand a haplogroup without looking at all its parts, the same that the blind man could not figure out the elephant by just gouging its trunk/tail/leg. And while the blind men of the tale were justified by the limits that blindness (and the narrator) imposed on them, you do not have such limitations so it is an error on your side.
A most grave and conscious error and why I accuse you of pseudoscience.
"consider ALL basal sublineages and not some arbitrarily chosen".
ReplyDeleteMaju. It is you who is not considering ALL the basal lineages. Remember A? And N9/Y?
"Unlikely is not the same as impossible".
True. But you're claiming the 'nearly impossible'.
"The same reasoning applies for the SA and WEA branches"
Exactly. And I have given them due consideration. The same reasoning aplies to them all. Branches with long tails are very likely to have remained in the one isolated region for some time, rather than having migrated huge distances as minor representatives within larger populations.
"It is you who is not considering ALL the basal lineages. Remember A? And N9/Y?"
ReplyDeleteYour insistance on those two lineages (out of 12) is what I call "arbitrarily chosen".
I am considering them all the time but they are just two out of 12: 1/6 of the overall picture.
"But you're claiming the 'nearly impossible'".
I don't see it that way and I don't see how would this change in your hypothesis. There's almost always some haplogroups that are distant from the overall origin of their parent haplogroup. From M and N in relation to L3 to X2g in relation to overall X2 (and so on).
So it does happen and rather often.
"Branches with long tails are very likely to have remained in the one isolated region for some time"...
That's a nonsense: those are clearly minor branches that have been bouncing around, they are not any useful reference. Except when you consider all subclades together, then they weight as any other.
Much more logical is to pay special attention to the branches with very short stems, as those expanded almost at the same time as (immediately after) the parent haplogroup did, so their track is more directly related if any.
"... rather than having migrated huge distances as minor representatives within larger populations".
There are so many possible stories about each haplogroup of that kind that we cannot tell them all in all our lives. Maybe it was part of a somewhat larger population that went almost extinct and was absorbed, maybe they were pushed around by more powerful bands till they found a place where nobody else had arrived, maybe they were 1/30 and by chance became, after many generations, 30/30. Maybe... there are infinite possible stories but I am not a novelist so I have to let your imagination flow...
Anyhow neither A nor N9 are so clearly detached from SE Asia: they are found all the way from SEA to NEA and the case is not very clear anymore for a NEA origin for them.
"Maybe it was part of a somewhat larger population that went almost extinct and was absorbed, maybe they were pushed around by more powerful bands till they found a place where nobody else had arrived, maybe they were 1/30 and by chance became, after many generations, 30/30. Maybe..."
ReplyDeleteMaybe, maybe, maybe. It's still most likely that they eventually expanded from the region they first became isolated in.
No. That's just one of the zillion possible scenarios. When the stem is long, the track is cold and you can't say much. Full stop.
ReplyDelete"That's just one of the zillion possible scenarios".
ReplyDeleteOK. Let's turn to your scenarios:
"Maybe it was part of a somewhat larger population that went almost extinct and was absorbed"
That doesn't shift them one metre from where they are found today.
"maybe they were pushed around by more powerful bands till they found a place where nobody else had arrived"
If they were being 'pushed around by more powerful bands' it's unlikely they'd be able to find a previously uninhabited region. Some haplogroups are today found in mountainous regions, for example, but they are usually survivors of a more widespread population thay has been reduced elsewhere. It's not that they entered that region on their own to escape persecution.
"maybe they were 1/30 and by chance became, after many generations, 30/30".
Again that is most likely to happen in an isolated population. And haplogroup that is 1/30 is far more likely to become extinct rather than take over. Unless it develops some huge advantage the remainder of the population fails to adopt.
"Anyhow neither A nor N9 are so clearly detached from SE Asia: they are found all the way from SEA to NEA and the case is not very clear anymore for a NEA origin for them"
But mainly derived haplogroups are found in SE Asia. So your comment is a bit like saying that, because Y-hap R is very common and diverse in New Zealand it must have originated there. Especially as many men who are obviously of indigenous phenotype have the haplogroup.
"those are clearly minor branches that have been bouncing around"
Surely if they had been 'bouncing around' to any extent they are much more likely to have become extinct.
"Let's turn to your scenarios"...
ReplyDeleteIt's not worth the effort because there are, as I said, a zillion possible cases. We only need one individual, maybe one small family, to trigger a founder effect where and when the conditions are good.
"... it's unlikely they'd be able to find a previously uninhabited region".
Notice that all these founder effects we are discussing here are in the last "frontier" areas of the Eurasian expansion: North and West Asia. Those frontiers allowed for founder effects that were impossible (or most difficult) elsewhere.
My whole understanding of Eurasian colonization is one of expansion followed by the "closure" of areas and routes by mere demographic pressure, so in this stage only the remote and cold NE Asia (north of Beijing) and the Neanderthal territory of West Eurasia remained open for new founder effects. These two processes seem to have happened at about the same time, maybe indicating a strong pressure from the already colonized areas of the Tropical and middle East Asian zones pushing some peoples to find even more remote and difficult new places to live - in some cases at least with success.
"Some haplogroups are today found in mountainous regions, for example, but they are usually survivors of a more widespread population thay has been reduced elsewhere".
I'd have to assess each case on its own merits. They may well be localized founder effects too. If they are refuge areas, the lineage should be found, even if at lesser frequencies, all around, if they are only found in a particular mountain are it's probably a case of founder effect in the colonization of that particular area.
"And haplogroup that is 1/30 is far more likely to become extinct rather than take over".
Absolutely correct. But once in 30 cases (or whatever the precise figure) it will happen as I say. Getting three sixes in a three dice throw is most unlikely but happens anyhow. There were many many dices cast all around Eurasia, so some had to be triple sixes.
This is an important understanding when we are dealing with probability. And we are.
"But mainly derived haplogroups are found in SE Asia".
This is what I do not have so clear, because I have yet to see a comprehensive study on A and/or N9 for their whole area, certainly not in a language I can read.
If you have clear data, please provide it.
"So your comment is a bit like saying that, because Y-hap R is very common and diverse in New Zealand it must have originated there".
Nonsense: Modern migrations and genocides have nothing to do with what could happen in Paleolithic or almost any other pre-modern conditions. And you guys lack the key R2 half part of the issue, not to mention related Q and P*, so at most you'd be talking of R1.
"... they are much more likely to..."
Again it's just a matter of casting enough dices. Maybe 200 or 15,000 "private" haplogroups had to go extinct for X to make a founder effect in the Levant. We will never know the exact figures.
"There were many many dices cast all around Eurasia, so some had to be triple sixes".
ReplyDeleteAny other examples of a haplogroup with a long tail that can be shown to have come into a region as a minority within a wider population, and then having expanded on its own?
Ok, I'll try to list all mtDNA haplogroups with a long tail (5 or more SNPs) and then a meaningful expansion:
ReplyDeleteL0, L0a2, L0k, L0d, L0d1, L0d2
L1"6, L1b, L1c, L1c1a2, L1c2, L1c3
L5a, L5c
L2'3'4'6, L2, L2a, L2b'c, L2b, L2c,
L6
L4a, L4b2,
L3b, L3f1b, L3d, L3e1, L3e3, L3x, L3h1
N, N1b, W, N9b, A, R1, T, R7, F2, F3, R11, B4c1b1, B4f, B5b, R31a, P4a, U1, U2b, U2e, U3a, U4, U8a
M1, M51, M2b, M7a, M7b'd, M7b3, M8a1, C, M10, M11, M12, M13a, M28, M31a1, M41, M53, M73, D4b1a1, D4h3a
That's it I think (notwithstanding whatever errors I may have committed). Lots of them.
"I'll try to list all mtDNA haplogroups with a long tail (5 or more SNPs) and then a meaningful expansion"
ReplyDeleteThanks. Now show me which of these cannot be assumed to have been in the region it expanded from over the whole period of that long tail. All the L0s you mention expanded from somewhere in southeast or east Africa. Presumably they'd originated there. All the L1s look to have been long established in Central Africa, perhaps northeast of the Congo basin. The L5s look to be long established somewhere near, or in, Ethiopia. All the L2s in the Sahel region. L6 somewhere between Egypt and Ethioia. And so on.
Each of the haplogroups has expanded from somewhere within their main range today. They haven't migrated for huge distances before they expanded. And almost certainly none 'have come into a region as a minority within a wider population'.
I will provide an example with L3: if your nonsense theory would be correct, L3 would have to expand not from East Africa, where all basal lineages have short stems (1 CR mutation) but from SE Asia, where N seems to have coalesced.
ReplyDeleteAs I said that's a nonsense. Obviously N is the one that made the long journey (as L3n or pre-N).
Another example can be found in U, which, following your conjecture would have coalesced in Egypt (U1), something nobody will agree with.
"if your nonsense theory would be correct, L3 would have to expand not from East Africa, where all basal lineages have short stems (1 CR mutation) but from SE Asia, where N seems to have coalesced".
ReplyDeleteI don't know how you reach that conclusion. I was trying to point out that each of those haplogroups with long tails are actually no different from the ones with short tails. Haplogroups of both type have individually coalesced somewhere within their parent haplogroup's geographic range. The parent haplogroup had previously expanded and broken into regional varieties, exactly the same as have genera, species and subspecies. No haplogroup has moved far from where they coalesced from their parent haplogroup, before they have in turn expanded, no matter how long the tail.
So look at L3's parent haplogroup, L3'4. That's what tells us most about L3's origin. The length of tail is not relevant. And technically L3 in its wider sense did make it all the way to SE Asia.
"Another example can be found in U, which, following your conjecture would have coalesced in Egypt (U1), something nobody will agree with".
And for a very good reason. It's immediately ancestral haplogroup is R, which coalesced in SE Asia. But R expanded as far as Egypt, or somewhere near it. Perhaps just R's descendant U itself actually reached Egypt, amoung other places.
But back to the main point. It is very difficult, if not impossible, to place the origin of either N9/Y or A in SE Asia. And it makes sense that the development of an ice age would eliminate many haplogroups across Central Asia, dividing mtDNA N into eastern and western haplogroups. However it is again very difficult to explain the disappearance of N haplogroups through India.
"I don't know how you reach that conclusion".
ReplyDeleteHopefully I misunderstood you. But I understand that you are all the time saying that certain subclades of N, your capricious choice, must have been located all the time, prior to their consolidation where they are found at expansion. These clades you choose are almost always the ones with longer stems (= longer obscurity "private clade" periods).
So if you say that A means that N expanded from North Asia, then N means that L3 expanded from SE Asia (or maybe North Asia if we follow the thread).
Instead the opposite is true: the location of the expansion of N and, to lesser extent, M are less informative regarding L3 as a whole, than those clades with short stems, which are all the African ones.
"I was trying to point out that each of those haplogroups with long tails are actually no different from the ones with short tails".
They are different: the ones with short tails expanded soon after their parent haplogroups, while the ones with long stems did a lot later, time in which they may have wandered (or not).
"Haplogroups of both type have individually coalesced somewhere within their parent haplogroup's geographic range".
No. This is not a valid reasoning. Technically you are maybe correct because one mutation before N it is not yet N but something else (L3*, L3n, pre-N) but this does not apply if we are talking of the root haplogroup in its pure form (L3-root). Between L3 and N there are a series of private (or otherwise extinct now) clades which are distinct evolutionary steps:
L3-root
pre-N (1)
pre-N (2)
pre-N (3)
pre-N (4)
N (N-root)
In this sequence we know (I understand) the locations of L3-root and N-root but there are four intermediate steps which are totally unknown. Let's see:
pre-N (1) is parallel to all the African L3 basal clades (five in number)
pre-N (2) has no parallel other than two steps-derived L3 subclades, of which I can only find L3i. So maybe at that stage pre-N was in Arabia (or not, but in this case it makes good sense).
pre-N (3) is parallel to M, so maybe then pre-N was in South Asia.
pre-N (4) is the most difficult to compare, so I won't.
N-root finally was in SE Asia, we have deduced.
N did not coalesce in the L3 range... unless you consider the obscure pre-N (4) or the better know one-step derived M subclades to be that. N in fact coalesced half a world away from where L3 existed when it took shape as such distinct haplogroup.
"No haplogroup has moved far from where they coalesced from their parent haplogroup, before they have in turn expanded, no matter how long the tail"
ReplyDeleteThe stem matters because the parent lineage of N is not in this sense you say L3-root but pre-N(4). However we do not call such kind of long lost lineages 'haplogroups' but this is a mere convention, as the word 'haplogroup' means group of extant (present day) haploid lineages. If pre-N(4) existed in, say Bangla Desh or maybe even already SE Asia, then what you say is correct... in relation to pre-N(4).
But certainly N coalesced a long distance away from where L3 existed and there is absolutely no problem with that precisely because its stem is five CR mutations long: enough.
Somewhat more problematic is when stems are short and yet distances are long, as happens with, specially, mtDNA M (see again this thread's map). You have sub-haplogroups, just one mutation away, in such distant places as South Asia, Japan and New Guinea.
So what do we have to conclude? That M-root spanned all that range? Sure, technically yes or something quite similar. But that does not apply for the moment in which M began expanding for sure, which must have been from some more specific location and a more specific population. What we have to accept is that there are many sub-mutation steps, which cannot be analyzed, but are the real people who carried mtDNA M-root around, eventually (soon) coalescing in all those subclades.
For this reason one-step mutations are most informative of the range reached by a haplogroup before deriving into recognizable sub-haplogroups. And for this reason they are more important.
"So look at L3's parent haplogroup, L3'4. That's what tells us most about L3's origin".
ReplyDeleteNope. It's L3 and L4 which inform about L3'4. We don't know shit about L3'4 without looking at its daughter clades.
"And technically L3 in its wider sense did make it all the way to SE Asia".
But only very technically and very much in its wider sense. IMO you are word-playing here.
If we consider single-step lineages (which must have existed in the past), L3-root never made all the way to SE Asia, only its descendants N (maybe pre-N(4) already) and M (in the form of would-be or maybe even already formed daughter lineages) did. But never anyone with the pure L3 sequence arrived to SE Asia: if they left Africa at all, they only reached as far as Arabia most probably.
"It is very difficult, if not impossible, to place the origin of either N9/Y or A in SE Asia".
I'm not sure right now about where they coalesced but in any case what I am saying is that the wandered from N's origin to wherever they did. Obviously the first N person was one single person who surely lived in one single place. Then, she or one or several of her descendants (not yet mutated into something else) was quite successful in expanding. What I'm saying is that, in the case of N, these people lived essentially in SE Asia and there's where they expansion began.
For me A is not different than HV, as both are five-steps removed from N. Just that HV's ancestors left a more clear trail (R, R0), while A's ancestors slided silently to their similarly remote destination.
"And it makes sense that the development of an ice age would eliminate many haplogroups across Central Asia"...
Excuses, nothing but pathetic pretexts! Why would that happen when some haplogroups clearly expanded to that same direction?
"However it is again very difficult to explain the disappearance of N haplogroups through India".
They just never left much of a mark, except for the most dynamic lineage, probably the largest of that N subpopulation. But all other "Western N" clades did leave some noticeable mark, with only one exception: X.
The more we discuss the more convinced I am all these Western N represent a single back-migrant population, which could not expand much in South Asia (as it was already densely colonized by the M clan).
"We don't know shit about L3'4 without looking at its daughter clades".
ReplyDeleteSurely it's the other way round. Its daughter clades surely formed and then expanded in turn from where their parent haplogroup L3'4 got to.
"They just never left much of a mark, except for the most dynamic lineage, probably the largest of that N subpopulation".
That 'apparent' explanation is totally inadequate. R left plenty of descendants as it moved through India, yet its parent haplogropup N did not. Strange.
"The more we discuss the more convinced I am all these Western N represent a single back-migrant population, which could not expand much in South Asia (as it was already densely colonized by the M clan)"
But R managed to colonise India very effectively.
"Just that HV's ancestors left a more clear trail (R, R0), while A's ancestors slided silently to their similarly remote destination"
Leaving no trace along the way. Strange.
"L3-root never made all the way to SE Asia, only its descendants N (maybe pre-N(4) already)"
Presumably N*. Where it formed haplogroups N21, N22, N13, N14, S and O. And presumably R, possibly somewhere between N21/N22 and N13/N14/S/O.
"I'm not sure right now about where they coalesced but in any case what I am saying is that the wandered from N's origin to wherever they did".
Surely it was N* that wandered at first. Its daughter haplogroups wandered once they had formed in the separate regions N had expanded through beforehand.
"What I'm saying is that, in the case of N, these people lived essentially in SE Asia and there's where they expansion began".
I suppose it's possible to manipulate the evidence sufficiently to claim that haplogroups A and N9/Y may have originated in SE Asia. But you have an even harder job placing X, N1'5 N2/W's origin in SE Asia.
I realise you dismiss Wikipedia unless it agrees with you, but this is what it says about various N haplogroups:
Haplogroup A:
"Its subgroup A1 is found in northern and central Asia, while its subgroup A2 is found in Siberia and is also one of five mtDNA haplogroups found in the indigenous peoples of the Americas ... 7.5% of the Japanese belong to haplogroup A (mostly A4 and A5).[3] 2% of Turkish people belong to haplogroup A".
Nothing about SE Asia (although derived haplogroups are found there) and certainly nothing about India.
The three basal haplogroups of N9:
"Haplogroup N9 - found in Far East.[18]
Haplogroup N9a - East Asia, Southeast Asia and Central Asia.
Haplogroup N9b - found in Japan.
Haplogroup Y[23] - found especially among Nivkhs and Ainus, with a moderate frequency among Koreans, Mongols, Tungusic peoples, Koryaks, Itelmens, Chinese, Japanese, Tajiks, Island Southeast Asians (including Taiwanese aborigines), and some Turkic peoples[17]"
Just one branch of one subhaplogroup in SE Asia (N9a). So It's difficult to make the case for an SE Asian origin for these two haplogroups.
"But certainly N coalesced a long distance away from where L3 existed"
ReplyDeleteAren't you forgetting Haplogroup X? Not to mention N1'5 and N2/W. Why do you feel it's necessary to postulate huge distances?
Sorry. I missed another one.
ReplyDelete"L3-root
pre-N (1)
pre-N (2)
pre-N (3)
pre-N (4)
N (N-root)
In this sequence we know (I understand) the locations of L3-root and N-root but there are four intermediate steps which are totally unknown".
We know L3 is African, but we don't know the location of N's root. Why are you absolutely certain it did not coalesce just outside Africa?
"So maybe at that stage pre-N was in Arabia (or not, but in this case it makes good sense)".
And there's no evidence that it did not remain there for the whole series of mutations. Then it expanded.
"N-root finally was in SE Asia, we have deduced".
No we haven't. You have decided. With no evidence. In fact evidence to the contrary.
Terry: we know nothing of L3'4 without looking at its daughter clades. Nobody is L3'4(xL3,L4) anymore and we do not have a time machine to go back in time and make genetic tests to the Idaltu clan or whatever. All we know of parent haplogroups is because of descendant ones living today (or in some cases of aDNA research as well).
ReplyDeleteReconstructing where L3'4 existed depends on first reconstructing where L3 and L4 did exist and is independent of where horizontal relatives such as L6, L5, L2 and L1 exist.
The process is always from today's actual haplotypes to yesterday's inferred haplogroups, what ca be a pain to calculate in large scale but can be done as I demonstrated in the African reconstruction maps (notwithstanding possible errors in the process).
It is obvious that you have never looked at haplogroups this way so, I'd suggest you to start doing some reconstructions yourself. Reconstructing N's original location from modern data should not be that hard, so go ahead and do it.
Yah, take a map of Asia-Australasia and begin the task of joining the dots.
"R left plenty of descendants as it moved through India, yet its parent haplogropup N did not. Strange".
Exactly what happened in West Eurasia, where R left plenty of descendants (what's that? 80-90%) and N only a bunch of lesser haplogroups, such as N1 (the most important of all, and also the first to branch out, after R, in the Western group), X and W.
You are aggrandizing the importance of residual N. Most N is R through the World, excepted some random peripheral areas like Aboriginal Australia.
So again thanks for revealing to me (albeit if confusingly) the SE Asian origin of R, it saves ancient humankind a whole journey back there and helps to explain N much better.
"Presumably N*"
ReplyDeleteN* means N-other in present day. There are always some unclassified lineages (or just not tested for in a particular research) that are placed under that umpbrella.
N alone or, more specifically, N-root or something like that should be used instead when talking of past pre-evolved "pure" N.
"Surely it was N* that wandered at first. Its daughter haplogroups"...
N* has no daughter haplogroups. It's residual N-other of today (a paragroup). I insist. Itself is descendant from N-root just like all other haplogroups in that clade, including all within R.
"I suppose it's possible to manipulate the evidence sufficiently to claim that haplogroups A and N9/Y may have originated in SE Asia".
I am not even saying that. I am talking of N-root before all these clades arose. It is the same in regard to the four Australian N subclades and the two known Negrito ones, etc.
"I realise you dismiss Wikipedia unless it agrees with you"...
I do not. As former active wikipedian, I know well that some stuff is accurate and other is not so much, so I appreciate when it's well documented and can be double-checked. Double-checking Wikipedia is generally a good idea, specially if you're working with specialist data, as we are doing here.
"Just one branch of one subhaplogroup in SE Asia (N9a)"...
And Y2, what makes it 1.5 (out of 3). Also I am quite sure that when I decided in the past to place N9's origin in SE Asia instead of further to the North it was because N9b is also found there.
Anyhow, it won't make a big difference re. the origin of N as a whole, because Negrito and Australian haplogroups weight a lot (6/12), so you'd need to place all other 6 in Lappland or something like that to argue for an Altai origin.
The real problem for your hypothesis is that there are not enough basal clades of N in Siberia or near it. So unless you want to argue that all SE Asian and Oceanian basal N subclades are ill researched and in fact (conjecturally) are a single haplogroup, you cannot argument your model at all. That's the only chance your model has: that somehow it's demonstrated in the future that SE Asian/Oceanian clades are tied by a single umbilical cord, now missing. Only that would displace the origin of N sufficiently to the North and West as to support your conjecture.
"that somehow it's demonstrated in the future that SE Asian/Oceanian clades are tied by a single umbilical cord, now missing".
ReplyDeleteThey are. They're all N haplogroups.
"Only that would displace the origin of N sufficiently to the North and West as to support your conjecture".
I've never claimed N 'originated' in the north. Only that it passed that way on its journey to East Asia. It's very difficult to avoid the conclusion that it originated just outside Africa when you look at the evidence.
"Negrito and Australian haplogroups weight a lot (6/12)"
You cannot use that sort of 'logic' to determine the region of origin of any haplogroup. There are any number of reasons why diversity might survive in one region and become reduced in another.
"N alone or, more specifically, N-root or something like that should be used instead when talking of past pre-evolved 'pure' N".
OK. I'm talking of the last of those N-root haplogroups when I say, 'And there's no evidence that it did not remain there [in Arabia] for the whole series of mutations. Then it expanded'.
"we know nothing of L3'4 without looking at its daughter clades. Nobody is L3'4(xL3,L4) anymore"
OK. The region of origin for L3 is likely to be somewhere near where the haplogroups most closely related to it are found. In this particular case L4. Same with haplogroups M and N.
"It is obvious that you have never looked at haplogroups this way"
Maju. I have always looked at haplogroups this way.
"N only a bunch of lesser haplogroups, such as N1 (the most important of all, and also the first to branch out, after R, in the Western group), X and W".
Haplogroups that had been in the west in some form from the moment the pre-N (L3N?) haplogroup left Africa. There is no evidence at all that they made the journey from to SE Asia.
"So again thanks for revealing to me (albeit if confusingly) the SE Asian origin of R, it saves ancient humankind a whole journey back there and helps to explain N much better".
We obviously still have a little way to go before you will be able to see the SW Asian origin of M and N though.
"You cannot use that sort of 'logic' to determine the region of origin of any haplogroup".
ReplyDeleteThat's how it is.
"There are any number of reasons why diversity might survive in one region and become reduced in another".
I do not think so, only extinction does that. And in any case the now extinct hypothetical Arctic sublineages of N are not any better sort of evidence than the now extinct such lineages in Madagascar, Ceylon or the Moon. Dead people do testify, hypothetical vanished lineages do not weight. First go and find them.
Because they have even tested for aDNA... but it is not N, M, L3 nor anything "ours": it is H. erectus mtDNA what is there (Denisova).
So there is even a negative evidence against your rant. Now please shut up and think.
"OK. I'm talking of the last of those N-root haplogroups when I say, 'And there's no evidence that it did not remain there [in Arabia] for the whole series of mutations. Then it expanded'".
Not sure what you're talking about here because your terminology is sloppy. If for you anything between the L3 and the A nodes is "N", but R is not, for instance, then we have a problem of understanding.
"The region of origin for L3 is likely to be somewhere near where the haplogroups most closely related to it are found".
Not necessarily. And again N is a great example: as its origin is half a planet away from where L3, L4, and all Ls are found, excepting itself and M.
"Same with haplogroups M and N".
M and N are not more related to each other than to L3x for instance.
"Maju. I have always looked at haplogroups this way".
Then why you are saying the opposite? Why are you saying that the origin of haplogroups must be located based not on their descendants but their ancestors and sibling clades?
"Haplogroups that had been in the west in some form from the moment the pre-N (L3N?) haplogroup left Africa. There is no evidence at all that they made the journey from to SE Asia".
There is one BIG evidence tatooed on the DNA: ALL them have exactly ALL mutations defining N at the root. If pre-N (L3n) branched out as you suggest, Western clades would be pre-N not fully evolved N.
So they were together (in a single personal line) for 5 full "eras" (single-mutation time periods) and only then in a single "era", they scattered around fast (but thinly, R excepted). But there were still another five "eras" before A showed up, four for X, etc.
"We obviously still have a little way to go before you will be able to see the SW Asian origin of M and N though".
Little is not the word here: forever is the word.
It is you who are absolutely wrong in this.
"Little is not the word here: forever is the word".
ReplyDeleteYou argued exactly that whenever I tried to persuade you of mtDNA R's origin in SE Asia too. So we'll await further evidence.
"And again N is a great example: as its origin is half a planet away from where L3, L4, and all Ls are found"
There is no evidence for that statement at all. Just your belief.
"M and N are not more related to each other than to L3x for instance".
Exactly. So why would we expect them to have coalesced half a planet away from somewhere near where L3a, L3b'f, L3c'd'j, L3e'i'k'x and L3h coalesced?
"Why are you saying that the origin of haplogroups must be located based not on their descendants but their ancestors and sibling clades?"
Because their descendant could have moved far from where they originated. Any haplogroup's descendants provide very little evidence regarding its origin, although the haplogroup presumably usually coalesced in a region where its descendants are still found.
"If pre-N (L3n) branched out as you suggest, Western clades would be pre-N not fully evolved N".
Why so? Not if its expansion, once fully evolved, was rapid. Surely you should argue the same for mtDNA R. Its eastern branches should be less fully evolved if it originated in SE Asia.
"So they were together (in a single personal line) for 5 full 'eras' (single-mutation time periods) and only then in a single 'era', they scattered around fast (but thinly, R excepted)".
Is that really so unbelieveable? I find it hardly remarkable at all. And there's no evidence at all that mtDNA N accompanied R on its expansion. In fact the evidence seems against the connection.
"But there were still another five 'eras' before A showed up, four for X, etc".
Again. What's so remarkable about that? The individual haplogroups remained isolated in various regions that N had passed through during its rapid route to SE Asia. They then remained isolated in those regions for some time, giving each haplogroup a long tail, until they were eventually able to indulge in expansions of their own.
"If for you anything between the L3 and the A nodes is 'N'"
And I was talking about N, not A. Conveniently for your belief N has a tail of five mutations. This allows you to shift its origin anywhere around the world that you find convenient. So you have it moving across India to SE Asia, leaving no trace of its route. And then moving back through India, again leaving no trace of its route. Amazing. You use the same 'logic' for haplogroup A. You simply shift it around to fit your current belief. And you reckon I make things up!!!
"You argued exactly that whenever I tried to persuade you of mtDNA R's origin in SE Asia too. So we'll await further evidence".
ReplyDeleteI hate how you put this matter as some sort of crusade or jihad you are in. We need no preachers, thanks. What we need is curious flexible-minded people who can see all the viewpoints of any matter, so the best solution(s) are found.
I may have some fun with preachers for a while but eventually I do get bored. So open your mind to what others say or get lost.
"So why would we expect them to have coalesced half a planet away from somewhere near where L3a, L3b'f, L3c'd'j, L3e'i'k'x and L3h coalesced?"
No "expectations" involved, sir, just factual data: it has nothing to do with ancestors but descendants and descendants say: South and SE Asia with impressive clarity.
"Any haplogroup's descendants provide very little evidence regarding its origin"...
Ok, it's clear we do not agree in method. We cannot therefore agree in anything else.
I have been all the time working that way: from descendant to ascendant, from bottom to top. And there is no other way, because we have no data for the past ancestor nodes other than the descendants of today.
And in the past you have followed and cheered my method, so what's the problem now? That it does not fit with your doctrine. Sorry but happens a lot in science.
[re. N] "Why so? Not if its expansion, once fully evolved, was rapid".
Oh, but, according to you, they had no boats, so they could not paddle their way. So now we have the "rapid migration through Tarim Basin" or what?
Did they already have domestic camels? Or maybe they are like Santa with flying reindeer?
"Is that really so unbelieveable?"
It has very low likelihood. Occam rules against.
...
...
ReplyDelete"And there's no evidence at all that mtDNA N accompanied R on its expansion"
They are found together in almost all places. So it becomes a strong possibility.
"Again. What's so remarkable about that? The individual haplogroups remained isolated in various regions that N had passed through during its rapid route to SE Asia".
But, in spite of being such a fantastic route by such marvelous lands for human habitation (whoa, Siberia, better than Bahamas!), they did not expand until much later. Please... it's a total rant!
Do you even read what you write? Do you even bother criticizing your own conjectures?
"Conveniently for your belief N has a tail of five mutations".
Yes, so extremely conveniently that it cannot be just luck.
"This allows you to shift its origin anywhere around the world that you find convenient".
No. I follow the geography of descendants: the descendants of L3 for the previous node (L3) and the descendants of N for N itself. It cannot be anywhere in the world: it must be the two specific areas I am saying, I cannot arbitrarily move these: the descendants won't allow me.
"So you have it moving across India to SE Asia, leaving no trace of its route. And then moving back through India, again leaving no trace of its route. Amazing".
How is it more amazing than a conjectural Siberian route with even less traces, which does not even fit the geometry of the descendants?
And anyhow they did leave traces in South Asia in the migration back to the West. Not many but enough. I'm now even pondering if W (most fequently found in North Pakistan) could be of South Asian origin, what would place all N2 in South Asia until the expansion of W itself.
You always accuse me of "belief" and nonsense. This is absolutely tiresome and boring. But you never look at the facts yourself: the descendants define the ancestor node in all aspects, including geography. At least that's how it works for us, looking at the matter from present-day data.
That's how it is: the descendants dictate where the ancestor was. I do nothing but listening to them. If you could listen (instead of actively searching for unlikely twists that might in your imagination support your preconceptions) you would see it crystal clear.
I do not manipulate shit: the haplogroups just speak themselves.
"I hate how you put this matter as some sort of crusade or jihad you are in".
ReplyDeleteI'm trying to help people understand what actually happened, not what they prefer to believe happened.
"And in the past you have followed and cheered my method, so what's the problem now?"
Because you're not using the same method as you've used to trace European haplogroups, and which you are still doing a very good job of in your latest blogs.
"No 'expectations' involved, sir, just factual data"
What 'factual data'?
"They are found together in almost all places. So it becomes a strong possibility".
But not in India.
"And anyhow they did leave traces in South Asia in the migration back to the West. Not many but enough".
What traces?
"I'm now even pondering if W (most fequently found in North Pakistan) could be of South Asian origin, what would place all N2 in South Asia until the expansion of W itself".
No it wouldn't. N2 breaks into two haplogroups after four mutations: N2a and W. According to Wiki, 'Haplogroup N2a - small clade found in West Europe.[20] Haplogroup W[21] - found in Western Eurasia and South Asia[22]'. Nothing exclusively South Asian all. Just that you wish it to be so.
"You always accuse me of 'belief' and nonsense".
Well, well, well. And there's an example of your own 'belief and nonsense'.
"But you never look at the facts yourself: the descendants define the ancestor node in all aspects, including geography".
But just because one line of descendants is found in a particular region, say SE or South Asia, doesn't mean the haplogroup must have originated it that region. The region of early branches can give a very good idea but when you use downstream mutations to justify a particular belief your argument moves onto shakey ground.
"How is it more amazing than a conjectural Siberian route with even less traces, which does not even fit the geometry of the descendants?"
It fits exactly the 'geometry of the descendants' but you prefer to place the descendants elsewhere.
"But, in spite of being such a fantastic route by such marvelous lands for human habitation (whoa, Siberia, better than Bahamas!), they did not expand until much later".
Ever heard of 'climate change'?
"So now we have the 'rapid migration through Tarim Basin' or what?"
Presumably something like that.
"I'm trying to help people understand what actually happened"...
ReplyDeleteWhat you think, or believe, happened. Instead what I demand is to look at matters for what they are: to look at the data without any preconceptions. Can you do that?
I think you cannot.
"Because you're not using the same method as you've used to trace European haplogroups"...
I think I am using the same method all the time. Otherwise explain me the differences, please.
"What 'factual data'?"
Where the haplogroups and the subhaplogroups, and the sub-sub-sub..-haplogroups (f)actually exist.
"But not in India".
In India too.
"What traces?"
R, N2 (incl. W) and N5 (half of N1'5).
"According to Wiki, 'Haplogroup N2a - small clade found in West Europe.[20] Haplogroup W[21] - found in Western Eurasia and South Asia[22]".
Metspalu 2006 (supplement) seems to indicate N2a being South Asian but maybe it's found in both regions, as happens with W and other haplogroups.
Whatever the case, let's assume for a moment all three Western N(xR) haplogroups are not found in South Asia (which is not true but whatever), where is the evidence pointing to Central Asia/Siberia? Nowhere!
So both hypothetical routes would be on the same situation. You still cannot prove that N migrated back to West Asia via the North or that it stayed all the time in the Far West, while the rest of N spread ultra-rapidly eastwards (without boats, to make things even more difficult).
"But just because one line of descendants is found in a particular region, say SE or South Asia, doesn't mean the haplogroup must have originated it that region".
Exactly my point: it is all the lineages together which describe where the ancestor was (by quite simple geometry).
"It fits exactly the 'geometry of the descendants' but you prefer to place the descendants elsewhere".
It does not. Have you tried to plot the dots on a map yourself and then find their centroid? It falls in SE Asia for N, there's no other solution and moving a few clades a bit here or there in Asia does not change that substantially because most of the weight is in SE Asia and Australia.
The only way that could happen would be if several of the SEA/Australasia clades would be joined into a larger N subhaplogroup, what would reduced their overall weight. But right now we have 7/12 clades pulling strongly towards SE Asia. And the rest do not pull towards West or Central Siberia anyhow but towards NEA and WEA. There's no subarctic diversity so early: it formed only at later times, at more downstream levels.
"Ever heard of 'climate change'?"
No matter: Bahamas will always be better than Siberia... unless they become a desert. Today we have some of the warmest conditions ever in the history of Humankind and Siberia is still a barren desert essentially, where human life is possible but extremely hard. And it's fucking cold!
"Instead what I demand is to look at matters for what they are: to look at the data without any preconceptions. Can you do that?"
ReplyDeleteYes I can. But can you? You have several times claimed that I fail to consider all the evidence when trying to explain the long tails of mtDNA haplogroups N, A and N9/Y. So let's look at some other haplogroups with long tails.
Within haplogroup N we find three other basal haplogroups with long tails: N13 has 13 mutations, N14 has 10 and N21 has 8. To apply your theory consistently would necessitate us assuming that the two Australian haplogroups really took their time moving from SE Asia to Australia while their tail grew, similar to the situation you claim for the original N migration from Africa to SE Asia. But the other two Australian basal N haplogroups, S and O, have respectively just 1 and 3 mutations in their tails. So, according to your theory, N13 and N14 must have been much later arrivals in Oz than were O and S. And the Malay haplogroup N21 has an 8 mutation tail. So, applying your theory consistently, we have to assume it must have moved very slowly from SE Asia to SE Asia!
When we turn to mtDNA R we find very long tails in several basal haplogroups. In the east we have R22 (in Indonesia, especally in the Lesser Sunda Islands) with a 10 mutation tail, and R23 (in Bali and Sumba) with an 18 mutation tail. These two haplogroups must also have taken a very long time to move from SE Asia to SE Asia! And what about R14 from New Guinea and the Nicobar Islands? A massive 23 mutation tail. That's enough to move to Africa and back again, possibly several times.
And in the west we have two more R haplogroups with long tails: R1 with 17 mutations and R3 with 19. According to your theory these haplogroups must have left SE Asia long after the other R haplogroups departed, and they moved extremely slowly west leaving no descendants as their tail developed.
In all the above cases isn't it much more likely that their long tail is a result of drift acting for some time on each separate haplogroup once it had become isolated somewhere within their ancestral haplogroup's geographic distribution? They each later embarked on their own individual expansion.
So isn't it extremely likely that we have the same general explanation for the tails of mtDNA haplogroups N, A and N9/Y, and even for the relatively short M tail? M and N each coalesced as a result of drift acting for some time on each separate haplogroup once it had become isolated somewhere within their ancestral haplogroup's geographic distribution, in this case L3.
And A and N9/Y each coalesced as a result of drift acting for some time on each separate haplogroup once it had become isolated somewhere within their ancestral haplogroup's geographic distribution. No need to postulate long migrations that left no descendants along the way while the tail developed.
Read my lips: my "theory" does not say lineages provided of a long stem traveled necessarily... it just enables them to do that more calmly than if they have a single-transition stem.
ReplyDeleteI'm not saying that stem means travel... I'm just saying it does not mean staying put in a shell as you claim.
Is this clear? Please respond YES/NO. If no explain your doubts.
"So, according to your theory, N13 and N14 must have been much later arrivals in Oz"...
ReplyDeleteThey might but they do not need to be so. The only thing long stems says is that the lineage survived in near-extinction conditions for long (for example as minority clade in a larger population or in an isolated clan in an island or even maybe that the lineage was violently pruned by a catastrophe of natural or human origin).
"In all the above cases isn't it much more likely that their long tail is a result of drift acting for some time on each separate haplogroup once it had become isolated somewhere within their ancestral haplogroup's geographic distribution?"
You want to keep each lineage in a box and that surely not possible. Drift of course was in action but did not need to act on a single isolated monophyletic "castes" (from Port. "casta": lineage, breed, race) but more probably in larger and more variegated real populations and population networks. There are just not enough isolated islands and deep gorge valleys to keep each lineage separated, specially when we know people moved around a lot in th early period of the Eurasian colonization.
"Is this clear? Please respond YES/NO. If no explain your doubts".
ReplyDeleteNo. It's not clear. You are inconsistent as to when you claim that 'stem means travel'. But you are consistently prepared to claim 'stem means travel' when it fits your belief, and, on the other hand, claim it doesn't mean travel when that perspective fits your belief. So how do you decide when to claim 'stem means travel'?
"Read my lips: my 'theory' does not say lineages provided of a long stem traveled necessarily..."
You're arguing with a fair degree of certainty that it does so in the case of mtDNA N, and pretty much so in the case of A. Why the inconsistency?
"for example as minority clade in a larger population"
You've already claimed elsewhere that a 'minority clade in a larger population' is unlikely to survive very long. And I agree.
"in an isolated clan in an island"
Not necessarily just on an island. All sorts of things can lead to population isolation.
"Drift of course was in action but did not need to act on a single isolated monophyletic 'castes' (from Port. 'casta': lineage, breed, race) but more probably in larger and more variegated real populations and population networks".
Quite. But if drift acts for any length of time on any 'larger and more variegated real populations and population network' it will very quickly eliminate all haplogroups but one.
"There are just not enough isolated islands and deep gorge valleys to keep each lineage separated, specially when we know people moved around a lot in th early period of the Eurasian colonization"
And isolation is so much more likely when the overall population is relatively small. Presumably that's why basal N (and Y-hap C) clades are so widely scattered.
Oh. By the way. Have you seem Wiki's entry on Y-hap C lately?
ReplyDeletehttp://en.wikipedia.org/wiki/Haplogroup_C_(Y-DNA)
Have a look at the map. It shows the haplogroup splitting into two. C5, C3 and C1 moving through Central Asia and C4 and C2 moving through India to SE Asia and Australia. I strongly suspect that there were no 'two routes' east though. We need another explanation. But going back to a comment you made the other day:
"Bahamas will always be better than Siberia... unless they become a desert. Today we have some of the warmest conditions ever in the history of Humankind and Siberia is still a barren desert essentially, where human life is possible but extremely hard. And it's fucking cold!"
Bahamas is batter, but if it's already full, or you can't get there anyway, you have to make do with Siberia.
"You are inconsistent as to when you claim that 'stem means travel'".
ReplyDeleteI do not claim that. Long stems only mean long coalescence times (as small "invisible" lineages), just that. They could have coalesced locally or they could have traveled or anything in between.
What means travel is when a haplogroup is found to have expanded far away from what is the logical reconstructed core of its parent haplogroup.
Is this clear?
"You're arguing with a fair degree of certainty that it does so in the case of mtDNA N, and pretty much so in the case of A. Why the inconsistency?"
No. I am arguing that N expanded far away from the logical origin of its parent lineage L3. I believe you agree with me in this. While a long stem helps to explain this (long time of migration) it is not a condition, as the migration can always be faster. You don't really need 5000 or 50,000 years to walk or sail from Arabia to Burma. Not counting modern obstacles such as borders, you can well do that in few years or even months.
But people was surely not in such a hurry in the Paleolithic and only migrated when they felt the need to do it (the details of why we can only speculate about).
"You've already claimed elsewhere that a 'minority clade in a larger population' is unlikely to survive very long".
Depends. If the population is large and growing the odds increase a lot. Drift acts only slowly (faster in smaller and decreasing or stagnated populations). It also depends on how much minoritary it is the haplogroup: it's not the same to be 1/1000 than to be 1/20, right?
Anyhow unlikely is not the same as impossible. Maybe hundreds or even thousands of other small lineages went extinct... but a few survived. Probability works that way: it's not mechanically straightforward, you know.
"All sorts of things can lead to population isolation".
Not sure if this is correct but certainly is difficult to explain why a clade would not expand for a long time and then suddenly do it massively, except if it happened to have arrived to a new "empty" land where to thrive. This applies specially to above average star-like expansions, which are all concentrated in "Greater Eurasia" (i.e. not in Africa) and must therefore, almost necessarily, reflect demographic expansions into virgin or quasi-virgin lands.
...
...
ReplyDelete"But if drift acts for any length of time on any 'larger and more variegated real populations and population network' it will very quickly eliminate all haplogroups but one".
But that assumes no population interactions, no expansions, no substructure... and even no stochastic randomness (or even real individual lives)... it is a good but highly theoretical lab model rather than a realistic one. There's no 1:1 map, reality always beats fiction (theory), etc.
Also I really disagree with your use of the phrase "very quickly". At the limit, when time tends to infinite, it unavoidably happens but time is always finite, and that's why the equation does not work so well in reality, even if it does reflect a real tendency.
If time=2500 generations (roughly 50,000 years), then it is still not infinite. And that is the largest figure I could came up with. "Very quickly" may mean what? Four generations? 50 generations?
Sure, the likelihood of "drift out" increases each generation that passes without expansion but it is never absolute. You can probably estimate how many other minor lineages had to go extinct, given X and Y circumstances, for one to survive, statistically speaking, but that's about it. You cannot ever confirm that any one lineage had necessarily to go extinct, much less when we know so little about the exact details. The odds may be against survival, the same they are against me winning lottery (assuming I'd buy a ticket, what I do not). But I can still win the big prize: there is a slim chance I do (and that's why people play lottery).
"And isolation is so much more likely when the overall population is relatively small. Presumably that's why basal N (and Y-hap C) clades are so widely scattered".
Population should have grown very fast in the conditions of a "virgin" Asia, so I think there must have been other factors at play, like multi-lineage expansions or second waves.
"Have a look at the map. It shows the haplogroup splitting into two. C5, C3 and C1 moving through Central Asia and C4 and C2 moving through India to SE Asia and Australia".
They must have changed the map in the last hours because now it shows the classical "because of C, then coastal migration" model. Central Asia is empty.
Checked the history and nope: the page has not been edited since September. You are suffering from some delusion.
"Bahamas is batter, but if it's already full, or you can't get there anyway, you have to make do with Siberia".
Fair enough (in the metaphoric realm). In fact Siberia was, in my understanding only colonized by AMHs after Tropical Asia was full. c. 50 Ka ago or later.
"Not sure if this is correct but certainly is difficult to explain why a clade would not expand for a long time and then suddenly do it massively"
ReplyDeleteThe development, or introduction, of new technology springs to my mind immediately.
"Long stems only mean long coalescence times (as small 'invisible' lineages), just that. They could have coalesced locally or they could have traveled or anything in between".
But they're far more likely to have coalesced locally.
"What means travel is when a haplogroup is found to have expanded far away from what is the logical reconstructed core of its parent haplogroup. Is this clear?"
Yes. Very clear. And, unlike you, I have nowhere claimed any haplogroup has 'expanded far away from what is the logical reconstructed core of its parent haplogroup'. Quote:
"I am arguing that N expanded far away from the logical origin of its parent lineage L3".
Why? It's not necessary to do postulate that N moved a huge distance from where it first emerged from Africa before it,in turn, expanded further.
"I believe you agree with me in this".
(continued)
"I believe you agree with me in this".
ReplyDeleteTry, for just a moment, to consider the possibility that mtDNA haplogroups M and N did actually first coalesce in separate regions somewhere near Africa. And try to look at the evidence with an open mind to see if it might be possible to untangle the data sufficiently to support such a possibility.
The tails show that the two haplogroups, pre-N and pre-M, were both subject to a considerable period of drift through either restricted population size or bottleneck before they were able to diversify, although M's population diversified rapidly once it had managed to burst into India after 3 mutations. It's quite possible that M1 remained for some time in the region where M had originally coalesed though. Members of M1 are found in Africa, but almost certainly a result of back migration. So that's M out of the way.
After 5 mutations N too broke free from the region where it had coalesced (Middle East or, very unlikely, SE Asia) and began to diversify, rapidly. However it's possible that the members of N who remained behind were not able to escape the established pressure from drift that had beset the original pre-N population. No SE Asia haplogroups demonstrates such a situation but X had 7 more mutations before it too began to diversify. N2/W experienced 5 mutations before it began its own population expansion, especially in Pakistan according to the information you supplied a few days ago. But by that time N5 had almost certainly already reached India because N1'5 split after just 1 mutation, and N5 is Indian. But neither N5 nor W moved very far through South Asia towards the east.
Haplogroup A underwent an 8 mutation long period of restricted population size after it had separated from the other N haplogroups. We'll leave open for now the question of where that population survived over that period.
But as soon as members of haplogroup N had reached the relatively benign habitat around the Yellow Sea the population expanded rapidly. After just 1 mutation haplogroup N9/Y split into N9a, N9b and Y. Interestingly, in his diagram of haplogroup N, Ian Logan still has haplogroups P and R basically descending from N9/Y.
Further south R began its own very rapid expansion after just 1 mutation. As did S in Australia. Haplogroup O expanded in Australia after 3 mutations. The other two Australian N haplogroups, N13 and N14, survived a long period of restricted population size (13 and 10 mutations respectively) and have not undergone any real population expansion. Nor have SE Asian haplogroups N21 and N22 with 8 and 7 mutations.
So that's it. All the mtDNA N haplogroups fitted snugly into a coherent sequence of expansion. No complicated two-way migrations leaving no descendants along the way. Nothing complicated at all in fact.
"In fact Siberia was, in my understanding only colonized by AMHs after Tropical Asia was full. c. 50 Ka ago or later".
But we don't know that for certain. And it was certainly colonized by some sort of humans long before 50 Ka.
"Try, for just a moment, to consider the possibility that mtDNA haplogroups M and N did actually first coalesce in separate regions somewhere near Africa".
ReplyDeleteIt does not work that way because then it would not have been M and N but some derived downstream clades the ones expanding. So pre-M and pre-N possibly did what you say but they did not quite expand yet.
"although M's population diversified rapidly once it had managed to burst into India after 3 mutations. It's quite possible that M1 remained for some time in the region where M had originally coalesed though".
Pre-M1 you mean? Possible but rather unlikely, it'd be easier for a pre-M clade to do that. Something like L3i, which is there to attest such kind of possibility, it seems, but is not the same line (a sister one).
"Ian Logan still has haplogroups P and R basically descending from N9/Y"
Logan's graphs are admittedly obsolete. Old stuff. Look at PhyloTree.
I'm done with this discussion, I think. You are not the least convincing to me. I do not think haplogroups the way you do: if two or more separate expansions happened in the line leading to N as you say we'd have Western pre-N and NEA pre-N as well, not N. That would be the evidence in favor of your hypothesis, which lacking that key evidence is nothing but wild speculation.
"I'm done with this discussion"
ReplyDeleteSo am I. Except for one thing:
"if two or more separate expansions happened in the line leading to N as you say we'd have Western pre-N and NEA pre-N as well, not N".
I have never claimed any more than one expansion of N. Just one expansion. It's your theory that has N moving backwards and forwards across half the world leaving no descendants anywhere along the way.
"It's your theory that has N moving backwards and forwards across half the world leaving no descendants anywhere along the way".
ReplyDeleteI think it's you who says that: you have N spawning subclades in West Asia, in Siberia and then again in SE Asia and possibly Australia too with your idea of large range of proto-haplogroups.
This is not possible. Even if the pre-N clade would have been hidden in some corner and then suddenly, with no preliminary track, you still claim that N-root spread all around Asia and Australia, yet without evolving to anything new (such as R or whatever). It is not just unbelievable but totally breaks the concept that we can track lineages at all, and that they were carried by normal people like you and me.
Because if, even before expansion, a haplogroup could be everywhere at the same time... we are before something more proper of quantum mechanics or relativistic physics, not about normal people living normal lives.
"I think it's you who says that"
ReplyDeleteNo it's not. Let me clarify what you believe happened. You claim that pre-N and pre-M emerged from Africa into the virgin habitat of SW Asia. They then traveled to the virgin habitat of India without leaving any descendants along the way. What's more pre-N then moved through India into SE Asia, again without leaving any descendants along the way. At which stage N diversified instantly into 12 different haplogroups, which expanded in various directions. Some of these haplogroups moved back through already occupied India, along with the N daughter haplogroup R. In that region R managed to leave many descendant haplogroups along the route but, yet again, N failed to leave any descendants. This whole scenario is surely very unlikely.
"you have N spawning subclades in West Asia, in Siberia and then again in SE Asia and possibly Australia too with your idea of large range of proto-haplogroups".
In contrast to your scenario I have N spawning various subclades as it moved (presumably rapidly) from Africa to Australia. Surely a much more likely scenario than yours.
"claim that N-root spread all around Asia and Australia, yet without evolving to anything new"
Again it is you who claim that situation. You have N moving huge distances around the world, often through virgin habitat, yet failing to leave any descendants behind. I totally accept the N-root left various haplogroups along its route, that eventually (or in some cases rapidly) gave rise to the surviving N-derived haplogroups. So this last comment of yours is completely stupid:
"if, even before expansion, a haplogroup could be everywhere at the same time... we are before something more proper of quantum mechanics or relativistic physics, not about normal people living normal lives".
I understand your reasoning but for me the scenario is most likely.
ReplyDeleteIn Arabia it was several lineages (L3i, L6, the various L0s...) because several sub-regions were at play (Yemen, Persian Gulf...) and Africa was just across the Red Sea (which for me is no real obstacle) bringing new people now and then, keeping the diversity somewhat high that way.
In South Asia insteadthe initial population was soon M-dominated, maybe with some pre-N and some other minor lineages but these eventually faded with drift. It'd be interesting that both of us looked in detail at the M subgroups because the expansion of N corresponds, IMO, not to the M stage but to two steps downstream (later in time).
So the question is not why N left no trace in SA but why it did expand so dynamically in SEA, when there were surely earlier arrivals within the dominant M clan which do not show such dynamism. I have no answer to this question but it's clear that N and R right after it, showed unusual dynamism, not just in SEA but at continental scale.
So we have a peculiar, highly mobile and expansive "tribe" dominated by the N haplogroup. Why was this? Without enough archaeological knowledge available I cannot tell.
"In contrast to your scenario I have N spawning various subclades as it moved (presumably rapidly) from Africa to Australia. Surely a much more likely scenario than yours".
Doesn't make sense because then the core expansion of N would be in West Asia, where the diversity is low (3/12) and then in NEA (as you defend the northern route), where diversity is even lower (2/12). Logically the center must be where diversity is high: in the SE (7/12 basal lineages, once we include R) and where the geometry of the whole haplogroup is centered (all agrees).
You put too much emphasis in L3 but ignore the fact that N does not show any sign of expansion for four full mutations/periods (at least 12,000 years, probably 16-20 millennia). And then suddenly it expands everywhere at the same time (according to you).
"You have N moving huge distances around the world, often through virgin habitat, yet failing to leave any descendants behind".
The people left descendants, no doubt but the lineage did not, as it was not dominant and was drifted out. Other co-migrant lineages did instead (M specially but several L clades too in Arabia).
What I think instead is of populations which had several lineages not always just one. Even in Papua, it's difficult to find such extreme homogeneity when we are talking of something bigger than a village: populations hold some diversity. They do tend to homogenization but this tendency is seldom fulfilled to its extreme.
Even Basques have two Y-DNA lineages (excluding erratics), even Native Americans have a bunch of them. Total homogenization in a population is rare.
"In South Asia instead the initial population was soon M-dominated, maybe with some pre-N and some other minor lineages but these eventually faded with drift".
ReplyDeleteSeems difficult to explain why one basal haplogroup would expand hugely while the other dissappeared entirely. As you pointed out:
"Even in Papua, it's difficult to find such extreme homogeneity when we are talking of something bigger than a village: populations hold some diversity".
And that diversity in Papua includes haplogroups of different origin, not just one.
"The people left descendants, no doubt but the lineage did not, as it was not dominant and was drifted out".
It which case it's absolutely stunning that N made it to SE Asia in the first place.
"Total homogenization in a population is rare".
Especially today. But I strongly suspect it was not too uncommon in the Paleolithic, although there has probably always been some wandering to and fro. But in the Paleolithic as any particular population moved into a new region, or developed a new way to exploit existing resources, their numbers would have expanded to the maximum sustainable. During the following period of relatively stable population numbers we get a bottleneck, in that haplogroups start to dissappear.
"Logically the center must be where diversity is high: in the SE (7/12 basal lineages, once we include R)"
But that diversity may be the product of major population expansion in the region as opposed to restricted population expansion, or bottlenecks, further north.
"So the question is not why N left no trace in SA but why it did expand so dynamically in SEA, when there were surely earlier arrivals within the dominant M clan which do not show such dynamism. I have no answer to this question but it's clear that N and R right after it, showed unusual dynamism, not just in SEA but at continental scale".
I have a theory but first we should do this:
"It'd be interesting that both of us looked in detail at the M subgroups because the expansion of N corresponds, IMO, not to the M stage but to two steps downstream (later in time)".
I agree. let's do it. M is very huge though, si it will be a big task.
"ignore the fact that N does not show any sign of expansion for four full mutations/periods (at least 12,000 years, probably 16-20 millennia). And then suddenly it expands everywhere at the same time (according to you)".
ReplyDeleteIt certainly expanded rapidly. But the interesting thing is that we can see the route it took, as long as we're prepared to concede it may not have moved far from Africa before it suddenly 'expanded everywhere'.
A haplogroup's long tail doesn't necessarily indicate any previous reduction in population numbers, just a population of restricted numbers over a period. Either situation could be described as 'a bottleneck', a restriction in population expansion.
"it's clear that N and R right after it, showed unusual dynamism"
Surely it is not usually the individual downstream haplogroups that migrate. For example let's look at mtDNA R, a haplogroup we both agree coalesced in SE Asia. Surely it is extremely unlikely the U, or even R0, were fully formed in SE Asia. It is much more likely that the group that departed from SE Asia contained a majority of what we might call, for convenience, mtDNA R*. Once this R* population had expanded U coalesced in SW Asia and replaced its parent potential haplogroup R*, through drift in the local population. Same with R0. Same with the other R haplogroups. In SE Asia itself R22 and R23 replaced their parent haplogroup R* along with many other pre-R22 and pre-R23 potential haplogroups. They have a very long tail but are found very near to where R itself coalesced. So their tail did not develope during a long migration. The same process presumably holds for mtDNA N, and M. I'll get back to you on M when I have time.
There are no pre-N anywhere, Terry. Let's accept the fact that N spread only AFTER M did. That applies specially to South Asia, where M's expansion happened first of all.
ReplyDeleteWas pre-N there? Possibly. How common was it? Not common enough to leave a mark.
So guess we have to accept that M was in South Asia when pre-N was still just part of a small population somewhere.
How did pre-N arrive to SE Asia? We cannot know but the coastal route is at least as valid as working hypothesis as any other, if not more.
"... as long as we're prepared to concede it may not have moved far from Africa before it suddenly 'expanded everywhere'".
I am definitively "not prepared to accept" that. For me N shows very, extremely clear, unmistakable signs of expansion in SE Asia. Full stop.
"That applies specially to South Asia, where M's expansion happened first of all".
ReplyDeleteI agree.
"So guess we have to accept that M was in South Asia when pre-N was still just part of a small population somewhere".
True.
"How did pre-N arrive to SE Asia? We cannot know but the coastal route is at least as valid as working hypothesis as any other, if not more".
Very unlikely if M had already filled South Asia.
"For me N shows very, extremely clear, unmistakable signs of expansion in SE Asia. Full stop".
But that still leaves the problem of how did N get there in the first place?
Turning to mtDNA hapogroup M. Certainly it could be said that 'suddenly it expands everywhere at the same time'. Basal haplogroups through South, East and Southeast Asia, and even to Australia and New Guinea. I make it 45 basal haplogroups in all.
ReplyDeleteI've no idea where 11 of them are from (M17, M25, M73, M72, M71, M60, M53, M52'58, M50, M49, M47 and M48) although I suspect that the majority are from Eastern India and Bangla Desh. From my notes (which are far from accurate) I make it 13 in South Asia, 7 in East Asia, 4 in SE Asia, 3 in Melanesia/New Guinea, 3 in Australia, 2 in the Andamans and 1 in Southwest Asia.
Two of the Australian haplogroups (M14 and M15) have a long tail and have not diversified at all. They could be the remanant of a wider population, similar to several of the SE Asian N haplogroups. Therefore they could have arrived in Australia at any time, especially seeing that they are confined to the northwest as far as I'm aware.
The New Guinea haplogroups present a dilemma. It seems humans didn't reach Australia until around 50,000 years ago, and the evidence for New Guinea is that arrival there was even more recent. So if we're to take the haplogroup mutation evidence at face value we have to assume humans arrived in New Guinea virtually simultaneously with M's expansion. That seems impossible if that expansion was immediately post Toba. Or perhaps M's expansion is much more recent, which would conflict with the mere 3 mutation tail at its base. What is the solution? I suspect that there are basal connections not yet discovered between several of the M haplogroups.
I also suspect it will be most productive if you first consider the M haplogropups that have diversified considerably, and then work back to try to work out their individual centres of expansion. Haplogroups M2, M4''64, M5 and M13'46'51 from India; D, M7, M8, M12'G and M13'46'61 from East Asia; M9 from SE Asia and M1'51 from Southwast Asia seem to be good candidates for such a study.
Look forward to any post you might care to make regarding any of these haplogroups.
"But that still leaves the problem of how did N get there in the first place?"
ReplyDeleteWe are discussing in circles. This is the "UFO abduction" problem I have mentioned several times.
You fail to acknowledge that several, maybe many, less important linages were all the time around but were eventually drifted out. We do not know their phylogenies nor their names but they were probably there until they finally vanished. The relative richness of Arabian Ls is testimony of what they once were surely.
One of those was pre-N, just that this lineage got particularly lucky and found/carved a niche for itself as N and R.
"You fail to acknowledge that several, maybe many, less important linages were all the time around but were eventually drifted out".
ReplyDeleteYou mean the way Neanderthal haplogroups have been 'drifted out'? As have those of other pre-modern humans. The process has presumably been operating since before we evolved from Australopithecus.
"Logan's graphs are admittedly obsolete. Old stuff. Look at PhyloTree".
Then you'll be interested to hear what he wrote to me today:
"Hello Terry
Thank you for your note.
Your point is well made - but I am not sure that is a clear answer as you have to take one stance or
another.
'Phylotree' chooses to keep '12705 & 16223' as a pair separating the R & N points.
But this means that there has to 'back-mutation' of 16223 for the sequences in haplogroup Y.
OK, this works; and the diagrams at www.phylotree.org follow this.
But, in my diagram I try to see if there is another possibility.
So if 12705 and 16223 can be split - then Haplogroup Y come can after the initial 16223 position
and before the later 12705
i.e. the Y sequences have the mutation 12705 (but not the mutation at 16223 )
And, this is what is seen - but in this instance there is the need also to include another
occurrence of the mutation 5417 in the Y stem.
I have a general dislike of 'back mutation' situations as mutations do not have any memory, so I
prefer the second view.
I do agree that my old diagram is not quite correct as it puts N9 in the wrong place, but I don't
think this really changes your point.
Let me know what you make of this.
Best wishes
Ian"
So there you have it. Not actually 'obsolete or Old stuff' at all.
What Ian Logan says is interesting indeed and I appreciate that you made that research effort. But it implies giving more importance to a mutation in the properly named Hyper-Variable Region I than to coding region mutation at locus 5417.
ReplyDeleteSure: it is a choice but I understand that the choice of prioritizing the mutation at 5417 is a logical one because mutations in the coding region are much rarer for the same locus (notice that the coding region is much larger so overall they maybe more).
There are other cases of mutations at 16223, as happens often with HVS loci (and that's why HVS sequencing is so imprecise and should be extended to at least relevant regions of the coding region). For example the same CRS-wise mutation is found in: X2h, M1a3, M7b3, M7c3b, M27b, Q1a'b'c, M39a2, D4c1, D4g2a1a, D4m1, D5c1, L3x2a, L0b, L0k2, L0d1a, L1c1a1 and L2d.
Inversely, there are further mutations at 16223 (back-mutations or mutations into a third state of the four possible ones?) within R at B5b1b and U4a2b.
I do not understand the use that Logan makes of the concept "back-mutation" in relation to Y. At best it is a parallel identical mutation, a coincidence (no "memory" anywhere). As you see these parallel mutations are common enough.
If you don't want to use the term obsolete, you can use "ill-maintained" or as Logan calls it himself: "old stuff". Certainly Logan's site was the only online reference before PhyloTree was launched last year but PhyloTree was very much necessary because Logan's page was far from the state-of-the-art and sometimes caused more confusion than solution.
Don't get me wrong: Logan's work is monumental and deserves praise but I understand that it overwhelmed him from some point on, and he seems to have lost interest or just could not keep it up (for instance, have you tried to search for haplogroup D?).
ReplyDeleteIn this sense PhyloTree, also a monumental work, is a clear improvement and a much needed update.
But I thank all them for their job, of course.
"I do not understand the use that Logan makes of the concept 'back-mutation' in relation to Y".
ReplyDeleteYes. He seems to have something backwards when he wrote, 'i.e. the Y sequences have the mutation 12705 (but not the mutation at 16223 )'. What he seems to mean is that the 'Y sequences have the mutation 16223 (but not the mutation at 12705)'.
"But it implies giving more importance to a mutation in the properly named Hyper-Variable Region I than to coding region mutation at locus 5417".
True. But what he finds surprising (I think) is that the mutation has happened at the root of two separate related clades (Y and R) at around the same time. However Ian still has a problem in that N9a and N9b do not have the 16223 mutation anyway. As I see it (if his tree is valid) is that both N9a and N9b must have lost the 16223 mutation. But 16223 is just one of many mutations in the Y tail.
"for instance, have you tried to search for haplogroup D?"
No I haven't, but that haplogroup may prove to be particularly interesting. It spreads in the east soon after it forms so should give a good idea of the route from South Asia to east Asia. Overall the M distribution tends to favour a more inland route rather than a coastal one. Most haplogroups appear to reach South China before they reach SE Asia, and (if we ignore the ones across Wallace's line for now) there are more of them north of what we might call SE Asia.
"But 16223 is just one of many mutations in the Y tail".
ReplyDeleteTrue but it's the only one in the same direction as in R, what is not uncommon, as I pointed put above.
If you make "Logan's choice", as I understand it, Y could be part of, let's call it, pre-R with a coding region mutation at 5417 identical to that of N9a'b (N9 by older nomenclature or if you accept the inclusion of Y). All the other mutations at Y's stem are particular of that lineage in either choice.
But "Logan's choice" is less parsimonious, albeit not too much if you consider HVR and CR mutations equivalent. Just that they are not, as I discussed above.
...
And don't bother searching at Logan's site for haplogroup D at Logan's site: it's just not listed at all. It was always frustrating for me not to find it.
Cheers.
"Most haplogroups appear to reach South China before they reach SE Asia"...
ReplyDeleteI consider South China part of SE Asia and what you call SE Asia (mainland) I call Indochina. But anyhow, this what you say makes some sense and is maybe suggestive of following the Brahmaputra and/or Irrawadi routes, as well as others (purely coastal and other rivers further east such as the Mekong, the Red River, the Pearl River, etc.
The clades that went southwards in most cases at least do not show any particular South China connection (rapid coastal migration? I think so). But some other clades may have gone to South China across Burma (I know you also dislike the idea but for me it's just another route and the Burmese highlands are definitively not impassable).
Of course a half-good Middle Paleolithic archaeological registry like the one there is in India, could help to located the inland routes more clearly. But following the South Asian big picture, I think that coastal and riverine routes must be the main ones.
But not necessarily the only ones. Non-Alpine mountains are no absolute barrier at all and even alpine type of mountains are always passable by the coasts. Here for instance the Pyrenees only become some sort of a barrier (probably impassable in the Ice Age) many dozens of kilometers away from the coast, elsewhere they are just rugged hills that a healthy person can walk through easily in a journey or even just hours, full with water and plentiful forests. Forests are rich indeed (boars, nuts, berries, fish, deer, mushrooms...) and plenty of wood to make fires, refuges and tools (rocks too). Resourceful hunter-gatherers would have a fine life in them.
Maybe a more diverse terrain is better but rugged terrain actually offers that kind of variety often, specially near the coast, where the weather is milder and you can complement land resources with sea ones.
My opinion anyhow.
"If you make 'Logan's choice', as I understand it, Y could be part of, let's call it, pre-R with a coding region mutation at 5417 identical to that of N9a'b"
ReplyDeleteYes. Either way he has the same mutation occurring in two separate lines. He's just told me he has redone his N diagram, but he still has the same problem:
http://www.ianlogan.co.uk/discussion/gifs/N_star_gif.htm
"The clades that went southwards in most cases at least do not show any particular South China connection (rapid coastal migration? I think so)".
They don't actually show any connection at all to any Ms anywhere, which I find very strange. I strongly suspect that is a result of inadequate research on the Oz/NG M-derived haplogroups. The main M haplogroup in SE Asia is M7. It survives as far out as the Moluccas and Borneo. SE Asian M8 survives no further southeast than mainland SE Asia, the Malay Peninsular and Vietnam. Likewise M21 and M22. That leaves just M7 as the only real island SE Asian haplogroup.
What about the trans-Wallacean haplogroups? The Melanesian haplogroups M27, M28 and M29 are not New Guinea haplogroups at all. They are confined to the islands immediately north and east of New Guinea, which again is very strange, although M29 descends from New Guinea Q. These three haplogroups may even be the product of a later movement across Wallace's Line.
I strongly suspect that the majority of the Oz/NG/Melanesian haplogroups will ultimately be shown to derive from M7.
"elsewhere they are just rugged hills that a healthy person can walk through easily in a journey or even just hours, full with water and plentiful forests".
I borrowed a book recently that claimed much of the Burman hill in the drier regions of that country was natural grassland. Easily negotiated by Paleolithic humans.
"Forests are rich indeed (boars, nuts, berries, fish, deer, mushrooms...) and plenty of wood to make fires, refuges and tools (rocks too). Resourceful hunter-gatherers would have a fine life in them".
It's becoming fairly evident that although heavily jungle-clad Borneo had been inhabited for a very long time life had been very difficult. The Austronesians arrived in a virtually uninhabited island.
I'm tired so I'll be concise:
ReplyDelete"I strongly suspect that the majority of the Oz/NG/Melanesian haplogroups will ultimately be shown to derive from M7".
When we know that for a fact, we will have reconsider whatever is needed. But at the moment it is just a conjecture (and mtDNA is relatively cheap to full-sequence and is pretty well surveyed nowadays, it seems)
"I borrowed a book recently that claimed much of the Burman hill in the drier regions of that country was natural grassland. Easily negotiated by Paleolithic humans".
Very interesting. What happened to your mud-infested uninhabitable highland jungles? ;)
"It's becoming fairly evident that although heavily jungle-clad Borneo had been inhabited for a very long time life had been very difficult. The Austronesians arrived in a virtually uninhabited island".
I do not understand why do you make that claim, specially when we have at least one people, the Mountain Dayaks or the Dayaks in general (unsure right now) that seem totally aboriginal (this I guess may be relative but at least pre-Austronesian). I would have to re-check right now but I think I remember that Borneo people clustered with Javans and some Sumatrans in the red "Austroasitic" component as well. Peninsular Malaysia with all its diversity was also part of the same Sundaland Peninsula in any case before the waters rose.
Anyhow, you could explain why your idea.
"What happened to your mud-infested uninhabitable highland jungles?"
ReplyDeleteThey're still the major part of the region.
"we have at least one people, the Mountain Dayaks or the Dayaks in general (unsure right now) that seem totally aboriginal"
I've linked to several papers in the past that made that claim but I haven't time to hunt them down just now. The majority of Y-haps in Borneo are Os of various kinds. Just a very small proportion of Cs, Fs and Ks, the most likely to be ancient indigenous. The mt DNA is less definite, but still could be a similar proportion. And there are certainly no groups that could be called 'Negrito'.
The Dayak are considered to be Austronesian in origin.
http://en.wikipedia.org/wiki/Dayak_people
From the link:
"Dayak languages are categorised as part of the Austronesian languages in Asia".
And:
"The consensus interpretation in modern anthropology is that nearly all indigenous peoples of South East Asia, including the Dayaks, are descendants of a larger Austronesian migration from Asia, thought to have settled in the South East Asian Archipelago some 3,000 years ago. The first populations spoke closely-related Austronesian languages, from which Dayak languages are traced".
The inclusion of SE Asia as a whole is obviuosly wrong as there are at least several pre-Austronesian groups in SE Asia, especially in mainland SE Asia.
"Peninsular Malaysia with all its diversity was also part of the same Sundaland Peninsula in any case before the waters rose".
I'm beginning to become convinced that the idea of a single Sundaland Peninsula has been exaggerated. It may have been so for just very short periods. The apparent separation of Peninsular Malay from Sumatra and Borneo is greater than we would expect if there was any sort of continuous Paleolithic connection.
"And there are certainly no groups that could be called 'Negrito'".
ReplyDeletePrecisely. Why are you so happily discarding all Y-DNA O (several clades) that Karafet claimed to be very clearly Paleolithic?
While I am not sure of the details, I am pretty sure that there is a lot of Paleolithic and pre-Austronesian Neolithic DNA still around.
I claim here the anti-copyright of this new aphorism: "Not everything Austronesian is necessarily Austronesian". (Google could not find an exact match, it's mine!!!) ;P
"http://en.wikipedia.org/wiki/Dayak_people"
"Do not believe all you read", they taught me. They could have added: "specially in Wikipedia".
I'll leave it at that: that they speak Austronesian languages does not make them Austronesians by blood (genetics). Languages and genes are not the same thing: compare Bob Marley with Mick Jagger...
Try revisiting the HUGO paper for instance.
"The apparent separation of Peninsular Malay from Sumatra and Borneo is greater than we would expect if there was any sort of continuous Paleolithic connection".
You mean in the genetic sense? I withhold my judgment but I see too many strange things: lineages claimed old that are most diverse, many genetically distinct populations, rather high diversity, contrasting "pure Austronesian" populations like the Mentawai and the Thoraya of Sulawesi, different from mainstream Indonesians, which show only partly that component, etc.
There was some kind of activity going on there before the Austronesian layer arrived. And if the Austronesian (or more properly Malayo-Polynesian) layer is more Filipino than Taiwanese, then the situation becomes quite complicated: who is what?
Austronesian is, specially in your discourse, an empty label. Is like discussing the genetic impact of Iberians in Latin America and then using the cultural tag Hispanic. Sure, it's a very smart way of increasing the numbers of Spanish or Portuguese... or Austronesians in our case, but from the viewpoint of genetics it does not change anything: biological ancestors are the ones who are whichever the language, religion, culture and identity.
So choose and define carefully which categories are you using and why they help to understand instead of blurring the picture by hyping the measure of their actual impact, specially genetic impact.
Consider please where cultural assimilation and/or some level of admixture may have happened. What evidence of the prior population we have still in the genes of modern Austronesian-speakers: are (some) Filipinos assimilated natives?, are they at the origin of the Malayo-Polynesians?, what impact had these populations in the Malay archipelago particularly?, again where do we find pure settling, where admixture, where assimilation?, etc.
Please.
"Consider please where cultural assimilation and/or some level of admixture may have happened".
ReplyDeleteI agree completely.
"So choose and define carefully which categories are you using and why they help to understand"
So I'll use the term 'Mongoloid' to encompass all later SE Asian arrivals, of which the Austronesians were one example, and 'pre-Mongoloid' for the Paleolithic inhabitants.
"Precisely".
Everywhere else in SE Asia Negritos are considered to be a remnant of the pre-Mongoloid SE Asians. The lack of Negritos in Borneo is a problem if claiming a substantial pre-Mongoloid substrate on the island.
"Why are you so happily discarding all Y-DNA O (several clades) that Karafet claimed to be very clearly Paleolithic?"
Because it's almost certainly incorrect. There was certainly a Neolithic genetic movement south into SE Asia, but if we don't attribute Y-hap O as being part of that we have nothing. O is much more likely to have originated in Central China.
"There was some kind of activity going on there before the Austronesian layer arrived".
I keep telling you that Austro-Asiatic is older in SE Asia than is Austronesian. On the mainland the Mon/Kmer, Thai/Kradai and Munda languages were part of the same expansion. I admit that German is convinced these languages have all existed in the region since the Paleolithic, but he is almost certainly incorrect.
"And if the Austronesian (or more properly Malayo-Polynesian) layer is more Filipino than Taiwanese, then the situation becomes quite complicated: who is what?"
I agree completely that the situation is quite complicated. But I doubt you would get any archeologist or paleoanthropologist to say the Dayak were anything other than Austronesian arrivals in Borneo.
"contrasting 'pure Austronesian' populations like the Mentawai and the Thoraya of Sulawesi, different from mainstream Indonesians, which show only partly that component, etc".
It's impossible to claim particular haplogroups as belongiing to specific cultural groups. That's a mistake many people make. Dienekes blog is full of people making such claims. All three O Y-haps are associated in specific regions with the Austronesian expansion. Same with mtDNA although it is difficult in that case to separate the ancient lines from incoming ones.
Well, it's clear (and thanks for clarifying) that you holds some assumptions I just cannot share.
ReplyDeleteOf course Negritos do look like a most old layer but I would expect the "proto-Mongoloid" (surely not yet the well defined type) one to be spread just after them, still deep in the Paleolithic and coincident with the spread of Y-DNA O all around.
I also think that Karafet is essentially correct in her assessment of Y-DNA in the area and that many clades of O and even O3 are deeply Paleolithic.
"I doubt you would get any archeologist or paleoanthropologist to say the Dayak were anything other than Austronesian arrivals in Borneo".
Arrivals or absorbed? We are talking genetics here, not just culture.
And anyhow there are two types of Dayaks who are genetically distinct from each other, in the HUGO paper they are described as Dayaks (Sea Dayaks, some 60% Austronesian apparently, the rest being mostly "Austroasiatic") and Bidayuh (Land Dayaks), who are at best 20-25% Austronesian by blood, displaying instead a 60% or more of a unique component (white).
These Land Dayaks, as well as the also distinct Proto-Malay of Malaysia seem to represent a different type of Paleolithic substrate to that of Negritos.
The Bidayuh look standard Mongoloid to me, while the Proto-Malay are more ambiguous maybe but still fit in. Yet I think they represent a Paleolithic element, along with the various Negrito groups.
"It's impossible to claim particular haplogroups as belonging to specific cultural groups".
I'm not discussing cultural groups here: I am discussing genetics and population replacement (or lack of it thereof).
If the Austronesian identity migrated without or with little genetic impact is a different case than if it did with almost total genetic impact (population replacement or colonization of virgin lands). I demand to be very clear of which layer(s) one means when saying "Austronesian" in these genetic debates and why.
It is important to determine how much pre-Austronesian blood is in modern Austronesians and how much is a direct arrival from Taiwan or wherever else. Your discourse clearly avoids this crucial matter and assumes that language/cultural identity means genetic replacement, what is clearly untrue.
"many clades of O and even O3 are deeply Paleolithic".
ReplyDeleteI agree O is 'deeply Paleolithic'. Just not deeply Paleolithic in SE Asia.
"Well, it's clear (and thanks for clarifying) that you holds some assumptions I just cannot share".
There has obviously been 'genetic replacement' in SE Asia, although many people seem determined to avoid having to face up to the fact.
Let's consider mtDNA M. I'm prepared to concede for the moment that it may have crossed the Bab al Mandab. It diversified incredibly once it had reached India, and expanded further east: into China south of about 30 degrees north, into SE Asia, and then out as far as New Guinea and Australia. So we would expect the people through the region to have all looked much the same at first. And in fact many people do see a modern phenotypic connection between some Indian tribals and both Ethiopians and Papuans.
So it's quite possible that there was originally a common phenotype stretching across South Asia from Ethiopia in the west to New Guinea in the east, and even to Australia. But the phenotype has vanished from South China and SE Asia for some reason. It has been replaced by a phenotype that looks like a hybrid between this original Ethiopian/Indian tribals/Papuan, pre-Mongoloid, phenotype and some sort of a Mongoloid phenotype. A phenotype that is far more pronounced in Northeast Asia.
It is very easy to conclude that this Northeast Asian population must have moved south at some time and interbred with the original South Chinese/SE Asia population.
Interestingly when we turn to Northeast Asia we find another apparently pre-Mongoloid phenotype: The Ainu of Northern Japan and Southern Sakhalin. Some people even see a phenotypic connection between the Ainu and the Australian Aborigines. There is the C Y-hap to support such a connection. Again it's possible that any pre-existing connecting population has been diluted by the subsequent expansion of, first, the southern pre-Mongoloid people and then, subsequently, the Mongoloid people themselves. The Mongoloid phenotype may not have reached Japan in strength until just 2000 years ago with the Yayoi, although it's reasonable to assume that these Yayoi were simply the culmination of a long process.
"It is important to determine how much pre-Austronesian blood is in modern Austronesians and how much is a direct arrival from Taiwan or wherever else. Your discourse clearly avoids this crucial matter and assumes that language/cultural identity means genetic replacement, what is clearly untrue".
The amount of pre-Austronesian blood depends on where in the Austronesian-speaking region you're considering. I hope the above helps explain the situation.
Some more thoughts on Borneo.
ReplyDeleteWe know there were people on Palawan at an early date, and Palawan has often been part of Borneo. And we know people on Palawan were eating taro at an early date. As I read somewhere recently, that tells us the people were hungry. Taro requires a great deal of preparation before it is safe to eat. So it is likely that Borneo was sparsely settled, at best, until the Austronesians arrived with their new technologies.
On the other hand we also know that during the Hoabinhian there was contact between Sumatra, Vietnam, and possibly the Philippines and even New Giuinea to some extent. Surprisingly there is no evidence for Hoabinhian influence in Borneo.
According to one of the papers German linked to recently it seems to have been the ultimate contact between the Philippines and Taiwan that lit the Austronesian fuse. It would be surprising if Borneo had never been part of any of the preceding movements. However contact from Sumatra to the Philippines may have been via Java, Sulawesi and Halmahera rather than via Borneo.
I'm sure that if we can get a handle on the gentic history of Borneo we will be able to work out exactly where mtDNA haplogroup R began its expansion from. The only pre-R haplogroup in Borneo (as far as I'm aware) is M7. But it is quite a widespread haplogroup so we need to look at which M7 is actually found in Borneo. It could have entered with R-derived either B5 or F1, or both at the same time.
"And in fact many people do see a modern phenotypic connection between some Indian tribals and both Ethiopians and Papuans".
ReplyDeleteI do not. All those groups are within what I call the Caucaso-Australoid continuum. Occasionally some Western Pygmies may have Papuan-like facial phenotypes but it's certainly not any pan-African or Ethiopian trait. It's more like the equally striking "coincidence" between Khoisan epicanthic fold (plus cheeks) and the East Asian one.
In any case, seen how difficult is to conciliate phenotype and genetics (there can well be epigenetic issues there or be described by a very small group of genes, selected by lookalike "identity selection"), I prefer to totally ignore the matter of phenotype, because it is more misleading than revealing so far.
"I agree O is 'deeply Paleolithic'. Just not deeply Paleolithic in SE Asia".
It's SEA what I am talking about. And for me O (and each subclade except arguably O2) has greatest diversity there.
"There has obviously been 'genetic replacement' in SE Asia, although many people seem determined to avoid having to face up to the fact".
I don't mind but show me clearly where and where from and in which amounts. I understand that this replacement is been greatly exaggerated.
"It is very easy to conclude that this Northeast Asian population must have moved south at some time and interbred with the original South Chinese/SE Asia population".
That idea does not even have archaeological support, much less genetic one. It's an old white man's myth: "Nordicism" in East Asia.
"The amount of pre-Austronesian blood depends on where in the Austronesian-speaking region you're considering. I hope the above helps explain the situation".
Without further clarifications this is Taiwan.
IF you think it's a good idea to introduce secondary gene pools, please describe them properly. For example, if you think that the peoples of Luzon were assimilated by early Austronesians, you may want to describe this secondary gene pool as secondary-Austronesian or Filipino-Austronesian or proto-Malayo-Polynesian, emphasizing the distinctiveness and not trying to hide it.
"That idea does not even have archaeological support"
ReplyDeleteYes it does. I've provided links in the past.
"I don't mind but show me clearly where and where from and in which amounts. I understand that this replacement is been greatly exaggerated".
There is certainly no evidence that O moved north.
"much less genetic one".
The spread of Y-hap O is pretty convincing evidence.
"if you think that the peoples of Luzon were assimilated by early Austronesians, you may want to describe this secondary gene pool as secondary-Austronesian or Filipino-Austronesian or proto-Malayo-Polynesian, emphasizing the distinctiveness and not trying to hide it".
The Austronesians as a whole have a varied genetic background, but the various combinations are ultimately simply a mixture of the same two kinds of people: southward moving East Asians and indigenous Papuans. A concept the German certainly couldn’t get his head around. This explanation will take a couple of posts, I'm sure, but I’ll try to explain the situation.
I certainly believe that the Austronesian language spread from Taiwan to the Philippines with Y-hap O1a. Perhaps the Philippines were largely uninhabited and that’s why the Austronesian language became so prominent there. I'm also fairly sure that mtDNA B4's expansion is Austronesian, although it has been pointed out that Taiwan B4 is a different clade to that of the Philippines. Although perhaps only 5-6000 years different. B4 is the main mtDNA haplogroup that Taiwan and the Philippines have in common although they do share B5, as a very small proportion of their total haplogroups. Both B4 and B5 are also found as far north as Japan, Korea, Northern China and Mongolia though. B4 is definitely associated with Austronesian-speaking people in the Pacific. In fact ultimately haplogroup B4a1a1a came to dominate Polynesian mtDNA haplogroups.
But B5 seems to most common on Sumatra and the Malay peninsular. And Java has B4 and B5. But Borneo has no B4 and just a little B5 as far as I know. So there's no Austronesian B4 on Borneo, yet the people universally speak Austronesian languages. How could that be so?
More than half of Borneo mtDNA is F1, which is a subclade of mtDNA R9. R9 is spread through South China and just makes it as far as the Malay Peninsular. So the Borneo F1 is probably the product of southward movement from there, or from Sumatra. It may belong to a pre-Austronesian substrate, most likely a product of the pre-Austronesian Austro-Asiatic expansion. But it may only have reached Borneo once the people had adopted the Austronesian language.
ReplyDeleteThe B5 of Borneo is particularly interesting. Haplogroup B appears to have been involved in the earliest, biggest, basal R haplogroup diversification. To me that suggests R and B originally expanded together, from the same place. Find B's origin and we have R's origin. B split immediately into B4, B5 and R24. And you could easily add R11 and B6 to the mix. Finding their centre of expansion is a challenge for you.
So, as Y-hap O1a and mtDNA B4 ventured out into the Philippines, and then into and around Wallacea they met and mixed with other people. Certainly Y-hap C in Southern Wallacea. C2a became the dominant haplogroup of the Austronesian-speaking Polynesians. O1a and B4 also met and mixed with populations containing Y-hap O2a, who had already come into SE Asia, perhaps with the Austro-Asiatic-speaking people. And populations containing O3, who had probably recently reached the most remote point of mainland SE Asia (the Malay Peninsular), having walked down through the hills of South China. And then they joined the Austronesians and moved through the islands with them. O3 has come to be the main surviving O haplogroup in the Polynesians.
Presumably several mtDNA haplogroups also joined the Austronesian shuffle. Austronesian languages came to dominate on all the islands of SE Asia and established a foothold in parts of the mainland coast. I suspect that the Austronesian-speaking people had improved their boating ability and many of the islands they reached had been uninhabited, or virtually so. Their more efficient use of the resources available led to their population growth.
I think I went through this at Dienekes, a couple of months ago:
ReplyDeleteC (C2?) and K (MNOPS) are Negrito clades. From what I recall also present in Melanesia and Wallacea and should correspond with the oldest crossing(s) of Wallace Line and the straits West of Philippines, maybe as early as the Middle Paleolithic.
O1a and O1a2 are specifically Austronesian clades, found in Taiwan Aborigines and Filipinos in great apportions.
Other O clades, mostly O3, must have other pre-Austronesian origin. In Philippines, O3* in particular is found among Southern Malay Filipinos and two Negrito groups, one of them from further North (Iraya). O3a3b seems related to North Filipinos but not Taiwan Aborigines.
Malay Filipinos seem divided in three clusters: North, West/Center and South. The Southern cluster looks the less Austronesian because of the high O3* (and some K and C); the Western cluster is dominated by O1a2, except the Hanunuo, who actually appear "Negrito" with all that K, while the Northern cluster is more diverse (O1a*, O1a2 and some "pre-Austronesian" O3a3b).
So I consider to be three layers in ISEA:
1. "Negrito" earliest layer (layer 1)
2. Pre-Austronesian "Mongoloid" layer 2
3. Austronesian layer (also "Mongoloid"): layer 3.
Confusing 2 and 3 is cause of all errors Austronesian.
"I've provided links in the past".
Nothing conclusive. You are yet to pinpoint the alleged "Northern" origin of the Mongoloid phenotype. The same that this phenotype is not found in SEA it is also not found elsewhere in all East Asia before Neolithic. We have skulls from South and North China, from Japan... and none are Mongoloid. And even in America (which you suggested once as the origin of the type) Ice Age skulls are not Mongoloid either.
"There is certainly no evidence that O moved north".
It's crystal clear for me. Each time a clade of O or O as whole is researched the South shows up as more diverse. As said the only exception could be O2 but even in this case, it's just balanced.
"The spread of Y-hap O is pretty convincing evidence".
For a South-North migration only.
"The Austronesians as a whole have a varied genetic background, but the various combinations are ultimately simply a mixture of the same two kinds of people: southward moving East Asians and indigenous Papuans".
Papuans (or otherwise non-Negrito Melanesians) are irrelevant except for Polynesia, which is a trivial region for our purposes.
This is not what I see: I see a pre-AN layer in ISEA that is not Negrito.
"So there's no Austronesian B4 on Borneo, yet the people universally speak Austronesian languages. How could that be so?"
Because languages are learned, not inherited biologically. Sounds quite basic to me.
"More than half of Borneo mtDNA is F1, which is a subclade of mtDNA R9. R9 is spread through South China and just makes it as far as the Malay Peninsular. So the Borneo F1 is probably the product of southward movement from there, or from Sumatra. It may belong to a pre-Austronesian substrate, most likely a product of the pre-Austronesian Austro-Asiatic expansion. But it may only have reached Borneo once the people had adopted the Austronesian language".
ReplyDeleteWhy? Why not Austronesian language being adopted as some minor but dynamic/dominant groups moved in. A lot of peoples, including Negritos, have adopted the Austronesian languages. I cannot pinpoint the exact processes but it does not seem too difficult to imagine, really.
"The B5 of Borneo is particularly interesting. Haplogroup B appears to have been involved in the earliest, biggest, basal R haplogroup diversification".
One of them, sure.
"To me that suggests R and B originally expanded together, from the same place. Find B's origin and we have R's origin. B split immediately into B4, B5 and R24. And you could easily add R11 and B6 to the mix. Finding their centre of expansion is a challenge for you".
Hypnosis: repeat with me: "it's a challenge for your..." :?
En fin, IMO B and R11 have the same SEA origin of R and N, very roughly. But feel free to correct me if I'm wrong.
"I suspect that the Austronesian-speaking people had improved their boating ability and many of the islands they reached had been uninhabited, or virtually so".
Borneo was not, nor anything in Sundaland, except probably the Mentawai islands. I think that you can pretty much reserve the "desert island" theory for the Pacific Ocean and little more.
"Confusing 2 and 3 is cause of all errors Austronesian".
ReplyDeleteYour 'Pre-Austronesian 'Mongoloid' layer 2' is most probably 'Austro-Asiatic', and also entered SE Asia from further north. So Os are all northern immigrants to SE Asia. O1a is Austronesian, O2a is the previous Austroasiatic and O3 is the most recent, arriving overland to the Malay Peninsular and then moving further east. O3 certainly became part of the Austronesian expansion because it's virtually the only O haplogroup in Austronesian-speaking people east of the Admiralty Islands.
"Nothing conclusive".
Only not conclusive because you efuse to accept what Peter Bellwood has written. His conclusions have never been seriously challenged by any scientist with knowledge about the region.
"We have skulls from South and North China, from Japan... and none are Mongoloid".
Isn't it quite possible that the 'Mongoloid' phenotype first appeared in Mongolia, or somewhere nearby?
"Each time a clade of O or O as whole is researched the South shows up as more diverse".
Such as in this paper?
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1226206/
Haven't you notice the lengths they go to to make the evidence fit the theory? Quote:
"To remove the influence of relatively recent population admixture, we constructed the STR network excluding the Tibeto-Burman, Altaic, Hmong-Mien, and southern Han populations".
They don't even consider the extremely likely possibility that those groups are simply the most recent southerly migratuons. And even after doctoring the evidence they still come up with :
"In general, the distribution of the O3-M122 haplotypes did not show distinctive divergence between southern and northern populations, with all the major subhaplogroups shared between them—except for O3-M7, which was observed only in the southern populations and therefore indicates a recent common ancestry of the O3-M122"
And:
"no significant differences were observed between SEAS and NEAS or among different language groups (data not shown)".
Doesn't sound like a Paleolithic separation to me.
Regarding southward migration:
"there are 14 Tibeto-Burman–speaking populations with a recorded history of migration from northern China ~3,000 years ago (Wang 1994)".
No Y-haplogroups were involved?
"I see a pre-AN layer in ISEA that is not Negrito".
It's Austro-Asiatic, and of course it's not Negrito. It's already a mixture of Negrito and Mongoloid.
"languages are learned, not inherited biologically. Sounds quite basic to me".
ReplyDeleteVery basic. But they have to be learned from someone, also quite basic.
"A lot of peoples, including Negritos, have adopted the Austronesian languages. I cannot pinpoint the exact processes but it does not seem too difficult to imagine, really".
But the Negritos are small minorities in a sea of Austronesian speakers. Quite obvious I would have thought.
"Why not Austronesian language being adopted as some minor but dynamic/dominant groups moved in".
What? You're proposing that some 'dynamic/dominant' group moved into a mountainous, jungle-clad island and imposed its language on the whole population?
"En fin, IMO B and R11 have the same SEA origin of R and N, very roughly. But feel free to correct me if I'm wrong".
But SE Asia is a pretty large and diverse region. We may be able to narrow the possibilities down considerably.
"Borneo was not, nor anything in Sundaland, except probably the Mentawai islands".
That statement is not in agreement with many specialists on the subject. Try reading Bellwood again, although this is not the paper where he looks at Borneo specifically:
http://epress.anu.edu.au/austronesians/austronesians/pdf/ch05.pdf
Page 5 has interesting comments regarding the settlement of western Indonesia. At the bottom of page 9 he starts his discussion on the pre-Austronesian populations in the region. On page 10 he mentions the pre-Austronesian Austro-Asiatic expansion.
You may be interested in this extract on the possible pre-Austronesian languages of Borneo:
ReplyDeletehttp://www.rogerblench.info/Archaeology%20data/SE%20Asia/Borneo/Hanoi%20IPPA%20text%20Blench%20submit%20circulation.pdf
I suspect they have the order of contact wrong though. The Austro-Asiatic substrate was on Sumatra, from where the acculturated population moved to Borneo. Evidently all the languages of Borneo have recognisable relations outside that island. They don't all diversify from just a single original dialect of Austronesian. This suggests multiple arrivals to the island.
"Your 'Pre-Austronesian 'Mongoloid' layer 2' is most probably 'Austro-Asiatic', and also entered SE Asia from further north".
ReplyDeleteThat's maybe part of it but Land Dayaks, proto-Malays and several other groups, including Wallaceans, show very clear Mongoloid features in spite of belonging excusively or mostly to clusters distinct from these two. So IMO there's a deeper "Mongoloid" layer.
In fact, for what we can see in DNA distribution we can make a broad parallel between Mongoloid features and Y-DNA O in ISEA at least. And O is at least 40 (IMO quite more).
It's easier if we are race-blind anyhow because appearence features do seem shallower than overall genetics (subject to social/sexual selection and may even be epigenetic in part).
"you efuse to accept what Peter Bellwood has written."
So I must recite "there's only one god Wallacea and Bellwood is its prophet"? And then what, cut a finger as proof of my blind faith?
"His conclusions have never been seriously challenged by any scientist with knowledge about the region".
Directly? Scientists avoid quarrel, which can damage their careers. Avoidance of direct confrontation is the rule, it seems to me.
I understand that there are many papers that challenge his ideas, just that they do not bother mentioning him.
"Isn't it quite possible that the 'Mongoloid' phenotype first appeared in Mongolia, or somewhere nearby?"
Not my concern, really. I'm interested in genetics, ancestry, not skin-deep traits, which should fit in and not be a conditionant, at least until their genetics are well understood.
When we get to understand what lays behind the mask of appearances in the genetic code, then we can address properly your race concerns.
"Haven't you notice the lengths they go to to make the evidence fit the theory? Quote:
"To remove the influence of relatively recent population admixture, we constructed the STR network excluding the Tibeto-Burman, Altaic, Hmong-Mien, and southern Han populations"".
You have quoted this paragraph before, and you have before been blind to the fact that both trees are presented: with and without said groups (fig. 6 A and B). And that if anything it could emphasize in some branches the role of SE Asians (South Han + Tibeto-Burmans) in the various trees. North Han never show up at the root in any case, so you always end up in the South. That's certainly true for Y-DNA O but also for other haplogroups.
"They don't even consider the extremely likely possibility that those groups are simply the most recent southerly migratuons".
Me neither, because if South Han were a recent arrival, we'd see them as less diverse than North Han and derived in all or most of their lineages. We do not, so they must be an older population, regardless of what language they speak now.
"Doesn't sound like a Paleolithic separation to me".
ReplyDeleteI do not think that there has been a clear cut separation anywhere in East Asia (from Lombok strait to at least the steppes and more likely the Arctic). Some island or otherwise isolated populations have remained more distinct (Ainu, Negritos) but in general the whole region seems to have been in fluid contact since early UP or earlier. It must be this fluid contact the reason behind your phenotype qualms (as phenotypes are product of convergence and not really expansion).
""there are 14 Tibeto-Burman–speaking populations with a recorded history of migration from northern China ~3,000 years ago (Wang 1994)".
No Y-haplogroups were involved?"
I really do not believe in the northern origin of TB. I think that TB may be related to Sichuan Epipaleolithic (and later Neolithic), and Sichuan is rather southernly. I also think that Sinitic may have diverged from TB in early Neolithic because this Sichuan Epipaleolithic and North China Neolithic seem related. Instead South China Neolithic could well be of Daic (+Austronesian?) linguistic character. It seems hard to tell about the Hmong-Mien origins but they also seem to be a local SEA group, maybe related to the Pearl River Corded Ware culture (distinct from the European one of the same name). The role of Daic and Hmong-Mien in South China Neolithic might well be the inverse one because they overlap very much.
"You're proposing that some 'dynamic/dominant' group moved into a mountainous, jungle-clad island and imposed its language on the whole population?"
ReplyDeleteYes. But more like they learned their language and dropped their ancestral ones gradually, within the context of trade, conquest, religious influence (Hinduism probably played some role in the expansion of Malay languages), etc.
There are many millennia for that to have happened in a criss-cross of indentitarian clashes and challenges of all types.
"But SE Asia is a pretty large and diverse region".
Not so extremely diverse, as we can see in all this discussion: they are interconnected very intensely.
On Bellwood:
ReplyDelete"An observation relevant for this question, one particularly intriguing in terms of its relevance for world prehistory, is that the general homeland regions of many of the major language families which have had long histories of association with agriculture seem to be geographically correlative with regions of primary
(i.e. indigenously-generated) agricultural origins (Bellwood 1990b, 1991, in press b). In the Old World such language families include Indo-European, (...) Afro-Asiatic, (...) Sino-Tibetan, Austroasiatic (...)"
I dispute this:
1. IE is not related to any primary Neolithic region, in fact they were very peripheral initially.
2. Afroasiatic expansion seems essentially pre-Neolithic. Only Semitic expansion can be safely associated to post-Neolithic events and again they appear to be a peripheral pastoralist group invading more consolidated "indigenous Neolithic" peoples.
3. Sinitic may well be related to North China Neolithic but Tibeto-Burman seems totally peripheral.
4. Austroasiatic is totally peripheral, as is Taiwan (and Philippines). Its expansion is not directly one of spread of "primary" Agriculture nor Taiwan was one of the "primary" Neolithic areas of the region at all. Their success seems more related to navigation and exploitation of sea resources than Neolithic proper.
While I can tentatively agree that SEA and ISEA early Neolithic (west of Wallace Line) was probably a matter of Austroasiatic speaking peoples, I suspect that there is more than just a mere AA demic expansion in all this and that we have to pay due attention to old distinct groups such as Proto-Malay and Land Dayaks (there are probably others). I am not even sure these only expanded in Neolithic but may well be of Paleolithic origin to at least some extent.
As for Blench's paper:
ReplyDeleteBut of course, Polynesia has never really been the problem; it is the large complex islands and archipelagos such as the Philippines, Borneo and Sulawesi that have to be explained. In recent years there has been a rising chorus of discontent from archaeologists who are increasingly claiming that the data doesn’t really fit the simple demographic expansion model.
So there seems to be nothing less than a whole "chorus" of experts raising questions against the overly simplistic Bellwood model. Ummm...
... the patterns of material culture in prehistory seem to point to earlier and more complex inter-island interactions than the Austronesian expansion model would seem to imply.
[Since 5000 BCE] ... a maritime culture was carrying trade goods around the region which would account for the similarities, without any the need to invoke demographic expansion.
... Austronesian languages in Borneo show borrowings from Austroasiatic languages.
The map at the conclusions suggests non-AA linguistic substrate in Sumatra specifically. It'd be therefore interesting to get wider sampling of West Indonesians and compare with neighboring peoples (Indochina, Wallacea, Philippines and Taiwan).
I think it can be summarized anyhow as:
1. Deepest Paleolithic layer (Negritos, earliest colonization).
2. Secondary Paleolithic layer (ill defined but almost certain, non-Negrito peoples with unknown languages).
3. Early Neolithic layer (AA)
4. Austronesian layer
We should not be able to cluster together any of the Paleolithic layers: the populations should tend to cluster on their own localized groups instead. Only the Neolithic layers should look homogeneous in the autosomal structure analysis.
BTW, I just stumbled upon the latest exercise by Dienekes on clustering skulls and we have some six or so different "Mongoloid" clusters: Amerindian (South America), East Asian (excluding NEA), Easter Island, Santa Cruz (North America), Buryat, Moriori/Maori and Inuit ("Eskimo").
ReplyDeleteThis is 6 out of 15 clusters in a global sample. Admittedly the large number of "Mongoloid" skulls may bias in favor of assigning more clusters to this heterogeneous group but still other extensively sampled areas as West Eurasia generate one single cluster (Norwegians and Egyptians are more identical than Buryats and Filipinos, for example), while the also heterogeneous group of "Australoids" behaves like "Mongoloids" (many distinct clusters) in spite of lower sampling.
In brief, the craniometry of Dienekes clearly shows that there is no one single Mongoloid type. However there is phenotype homogeneity in the majority population from Beijing (steppe frontier) to Wallace Line, including Japan (but not Ainu). I'd suggest to use the term "Sinoid" here for clarity, as it's obvious that Mongols are excluded.
"That's maybe part of it but Land Dayaks, proto-Malays and several other groups, including Wallaceans, show very clear Mongoloid features in spite of belonging excusively or mostly to clusters distinct from these two".
ReplyDeleteBecause they're a hybrid population. That's why they have formed their own cluster distinct from Chinese and Papuans.
"In fact, for what we can see in DNA distribution we can make a broad parallel between Mongoloid features and Y-DNA O in ISEA at least. And O is at least 40 (IMO quite more)".
And O almost certainly began its spread with the Chinese Neolithic, in spite of your continued claims otherwise. Several mtDNAs also look as though they too have come south with Y-hap O:
http://www.ebc.ee/EVOLUTSIOON/publications/Yao2002.pdf
Admittedly from 2002. From the distribution map, and the authors' statements, C/M8 appears to have an origin in the northern half of China, at least north of Shanghai. A, N9, G and Y also all look to be originally from north of Shanghai. Even M7's origin is probably no further south than Hong kong. Of course the R-derived haplogroups all show a southern origin and subsequent move north.
"you have before been blind to the fact that both trees are presented: with and without said groups (fig. 6 A and B)".
And removing them actually makes very little difference.
"because if South Han were a recent arrival, we'd see them as less diverse than North Han and derived in all or most of their lineages. We do not, so they must be an older population, regardless of what language they speak now".
Not true. The Southern Han are a mixed population, just as the SE Asians now are. Hence the southern diversity that shows up repeatedly.
"I do not think that there has been a clear cut separation anywhere in East Asia (from Lombok strait to at least the steppes and more likely the Arctic)".
No. We basically have a cline that stretches all the way. The cline is the result of prehistoric and historic to and fro movement. And that is why 'the craniometry of Dienekes clearly shows that there is no one single Mongoloid type'.
"I really do not believe in the northern origin of TB".
What? That's why its removal makes so little difference to the data in fig. 6 A and B?
"I also think that Sinitic may have diverged from TB in early Neolithic because this Sichuan Epipaleolithic and North China Neolithic seem related".
Ans is probably the source for the expansion of Y-hap O3.
"dropped their ancestral ones gradually, within the context of trade, conquest, religious influence"
Some inland Borneo groups have not been subject to any of those things.
"IE is not related to any primary Neolithic region, in fact they were very peripheral initially".
I agree that IE does not appear to ba at all associated with any agricultural expansion. That belief was part of the support for an Anatolian origin for IE, which I'm sure is incorrect.
"Sinitic may well be related to North China Neolithic but Tibeto-Burman seems totally peripheral".
But the two language groups are probably related. Therefore they have a common origin or were originally part of a language chain.
"Their success seems more related to navigation and exploitation of sea resources than Neolithic proper".
ReplyDeleteTrue. Improved boating technology.
"So there seems to be nothing less than a whole 'chorus' of experts raising questions against the overly simplistic Bellwood model. Ummm..."
Only over the 'simplicity' of it. Most biological processes are complicted so we could assume human migration have been the same.
"The map at the conclusions suggests non-AA linguistic substrate in Sumatra specifically".
Non-Austronesian rather than non-Austro-Asiatic isn't it? And I strongly suspect the movement was from Sumatra to Borneo, rather than the other way.
"It'd be therefore interesting to get wider sampling of West Indonesians and compare with neighboring peoples (Indochina, Wallacea, Philippines and Taiwan)".
Very much so. That's where Dienekes data set falls down.
"I think it can be summarized anyhow as"
I agree with your assesment.
"Secondary Paleolithic layer (ill defined but almost certain, non-Negrito peoples with unknown languages)".
Possibly related to Melanesians or Australian Aborigines.
"I'd suggest to use the term 'Sinoid' here for clarity, as it's obvious that Mongols are excluded".
OK. We have a Sinoid movement south starting during the early Chinese Neolithic. In fact the movement continues to this day. The recent problems in East Timor, and the current problems in Irian Jaya are just a continuation of this ongoing process.
1. Cluster homogeneity is NEVER sign of admixture. Recent admixture is ALWAYS found as two or more components. That's the main characteristic of Structure-like algorithms.
ReplyDeleteThe main exception may be when the intrusive component is shown and the aboriginal one is not found because it ranks too low in relation to the overall sample set. So maybe some of those Austronesian/Austroasiatic populations also hide their own aboriginal lesser clusters, which would only show up at deeper Ks or with a different sampling strategy.
"Because they're a hybrid population. That's why they have formed their own cluster distinct from Chinese and Papuans".
What?! "Hybrid" (not a word used in humans, really, nor appropriate for admixture within the same species) or rather admixed peoples, such as African-Americans or Mexicans, do not develop their own clusters: they remain showing that admixture very very deep into the k clustering.
Most of these populations do show admixture components but they also show their own uniqueness as dominant. You could argue maybe for inbreeding (accelerating the homogenization process) but never for admixture in such cases.
Also, as they show intrusive components (Austronesian and/or Austroasiatic typically), we can conclude, I understand, that the arrival of these intrusive components is more recent than the culmination of the homogenization (or inbreeding) process.
"And O almost certainly began its spread with the Chinese Neolithic, in spite of your continued claims otherwise".
ReplyDeletePlease!
"Several mtDNAs also look as though they too have come south with Y-hap O:
http://www.ebc.ee/EVOLUTSIOON/publications/Yao2002.pdf"
A 2002 paper on Han only. If all your point is to demonstrate that Han have their origins in North China, I can agree with that... but it's quite meaningless for the overall population structure and even pretty much secondary for Han themselves, who are defined by language and identity rather than blood (as happens with most nations).
"C/M8 appears to have an origin in the northern half of China"
Likely true. I have generally considered M8 (including CZ) to be one of the pioneers in the colonization of Mid and North East Asia.
"A, N9, G and Y also all look to be originally from north of Shanghai".
At this moment I have not clear the situation for A and N9 (incl. Y). G is related to a SEA haplogroup: M12, together they make up M12'G.
"Even M7's origin is probably no further south than Hong kong".
I've generally placed it not far from Japan and maybe in Japan itself.
"And removing them actually makes very little difference".
Exactly (well, it clarifies the structure a bit in fact). And that's why I do not understand why you keep insisting that the authors are somehow manipulating the facts re. O3. Well, I understand: intellectual laziness and inability to change your opinions according to evidence (and therefore the need to blame the evidence for your frustration and denounce it almost mechanically as biased, often without any grounds).
"The Southern Han are a mixed population, just as the SE Asians now are. Hence the southern diversity that shows up repeatedly".
That might be the case if we'd be talking of merely generic diversity but not when we are talking of BASAL DIVERSITY for nearly each haplogroup we can discern.
"We basically have a cline that stretches all the way. The cline is the result of prehistoric and historic to and fro movement".
I can agree with this, just that I'd simply remove the word "historic", as even the most recent flows, Austronesians and such, are prehistoric.
I'd emphasize anyhow the importance of the Paleolithic flows, because too many things, the very essentials of the regional structure, cannot be explained without them.
"And that is why 'the craniometry of Dienekes clearly shows that there is no one single Mongoloid type'".
No. It has nothing to do: continuous flows and continuous genetic cline should cause a homogeneous phenotype, as happens with the "Sinoid" cluster. The fact that the other "Mongoloids" are apart is because they have been at the margins of this flow since maybe 20,000 years ago or probably more.
"What?"
ReplyDeleteI repeat: I do not believe in the North China origin of Tibeto-Burman language family (nor its putative population carriers). Their origin seems to be at the Eastern edges of the Tibetan Plateau. In some papers they show up as a very distinct cluster from other East Asians, including Han.
"That's why its removal makes so little difference to the data in fig. 6 A and B?"
I don't think so. If O3 would have arrived from the North and TB peoples would have been carriers, their removal from the graphs would matter A LOT. I'm astonished at the lack of comprehension you are showing.
"Ans is probably the source for the expansion of Y-hap O3".
Please, I was talking languages: Sinitic and Tibeto-Burman. It is possible that the TB core area of Sichuan and the North China Neolithic (Sinitic) were related (while TBs remained Epipaleolithic still for a while). I am not making any statement about blood migrations, just language connections and archaeological cultural connections.
And anyhow there's no way to make O3 fit even the wildest narration of Sino-Tibetan expansion. First it has a southern centered structure and second Sino-Tibetans in general never conquered most of SEA.
It's crazy: you are so entrenched that you cannot see shit.
"Some inland Borneo groups have not been subject to any of those things".
They have been assimilated into the Austronesian language community somehow. Difficult to say because no one seems to sample them in any case. The Land Dayaks of the HUGO paper are from Sarawak, not far from Kuching.
"But the two language groups are probably related".
I'd say so but it looks like a very old relationship: a pre-Neolithic one maybe or at most from the earliest Neolithic. Still some people question the existence of a Sino-Tibetan family:
"A few scholars, most prominently Christopher Beckwith and Roy Andrew Miller, argue that Chinese is not related to Tibeto-Burman. They point to what they consider an absence of regular sound correspondences, an absence of reconstructable shared morphology[2], and evidence that much shared lexical material has been borrowed from Chinese into Tibeto-Burman".
Others instead argue that there are regular correspondences.
It seems only slightly more solid than Indo-Uralic.
"Non-Austronesian rather than non-Austro-Asiatic isn't it?"
ReplyDeleteSure, I stand corrected.
...
"Possibly related to Melanesians or Australian Aborigines".
No, no: Negritos, Australian Aborigines and Melanesians in general must have coalesced as various products of the first wave.
My second wave would include other non-Negrito, non-Melanesian, non-Australoid peoples. It would be made up of proto-Mongoloids, so to say, and include lineages such as Y-DNA O.
"OK. We have a Sinoid movement south starting during the early Chinese Neolithic".
Not what I think. I'd rather associate the Sinoid phenotype, at least in its basics to the expansion of Y-DNA O (in the early UP). In fact they overlap very well to the exclusion of North Asian/American lineages such as C, N and Q. The only exception would be Polynesians but they have so much Melanesian blood (haplogroup C2) that it's easy to explain why they do not cluster even with their Malay or Taiwanese cultural cousins.
"In fact the movement continues to this day. The recent problems in East Timor, and the current problems in Irian Jaya are just a continuation of this ongoing process".
Well... you are going a bit too far. The technologies of today allow for huge-scale genocides and colonizations, totally unthinkable before Modern Age.
A final spontaneous thought. The "Sinoid" craniometric homogeneity can be compared with the "Caucasoid" one. This last one we know (I do know) that is rooted in early UP expansions. In fact, Dienekes had Cro-Magnon (alpinoid) and Chancelade (nordo-med) clustering in that group, so it's at least of Gravettian age.
ReplyDeleteSo we can easily conclude that the "Sinoid" phenotype can also be from that date range (c. 40-30 Ka ago), which is the usual age assigned to Y-DNA O expansion.
This is ridiculous, but I'll carry on for now:
ReplyDelete"I do not believe in the North China origin of Tibeto-Burman language family (nor its putative population carriers). Their origin seems to be at the Eastern edges of the Tibetan Plateau".
Yes. Sichuan. Makes sense totally. A couple of Wiki links:
http://en.wikipedia.org/wiki/Yangshao_culture
http://en.wikipedia.org/wiki/List_of_Neolithic_cultures_of_China
You'll notice that the Chinese Neolithic had reached Sichuan by 3100 BC. And in an old textbook I have the author (Grahame Clark) has this to say(I have also quoted this before):
"Farming was introduced first into the lower and middle Yangtze basin, whence it spread into Szechwan and south into the coastal privinces from Chekiang to Kuangsi and beyond into southeast Asia"
My guess would be O3 in Szechwan and O2 into the coastal provinces. Fits the modern distribution very well.
"there's no way to make O3 fit even the wildest narration of Sino-Tibetan expansion".
O3 is often considered a marker of Tibeto-Burman expansion.
http://en.wikipedia.org/wiki/Haplogroup_O3_(Y-DNA)
From the link:
"Among all the populations of East and Southeast Asia, Haplogroup O3 is most closely associated with those that speak a Sinitic, Tibeto-Burman, or Hmong-Mien language. Haplogroup O3 comprises about 50% or more of the total Y-chromosome variation among the populations of each of these language families. The Sinitic and Tibeto-Burman language families are generally believed to be derived from a common Sino-Tibetan protolanguage, and most linguists place the homeland of the Sino-Tibetan language family somewhere in northern China".
"as even the most recent flows, Austronesians and such, are prehistoric".
But I think we could call the Han movement 'historic'.
"If O3 would have arrived from the North and TB peoples would have been carriers, their removal from the graphs would matter A LOT. I'm astonished at the lack of comprehension you are showing".
It would make very little difference if the TB people were just the later part of the same movement. In fact all the basal O haplogroups are widespread through both north and south, surely suggesting recent expansion. With the removal of the TB people we are left with just the downstream O-M7 as a completely southern haplogroup. In my notes I have this haplogroup described as Hmong-Mien and Malay, suggesting a connection between Tibeto-Burman and Hmong-Mien.
"I'd rather associate the Sinoid phenotype, at least in its basics to the expansion of Y-DNA O"
At least we agree on that. Our disagreement is merely over when that expansion occurred.
"So we can easily conclude that the "Sinoid" phenotype can also be from that date range (c. 40-30 Ka ago)"
Almost certainly so, but its presence in SE Asia is much more recent.
"No, no: Negritos, Australian Aborigines and Melanesians in general must have coalesced as various products of the first wave".
One minute you're quoting peoples' criticism of Bellwood for proposing too simple a process and here you are proposing just such a simple process.
"My second wave would include other non-Negrito, non-Melanesian, non-Australoid peoples".
Evidence?
"Not what I think".
I'll leave you free to think what you like.
"This is ridiculous, but I'll carry on for now".
ReplyDeleteDo not feel obliged, really. Nearly nobody reads the comments but those subscribed, you know.
"the Chinese Neolithic had reached Sichuan by 3100 BC"...
Do you mean that Sichuan remained out of "Chinese" Neolithic for at least 4000 years in spite of being just side by side with the most dynamic centers of "Chinese" Neolithic? These lazy Tibeto-Burmans! :)
"It would make very little difference if the TB people were just the later part of the same movement".
Damn, how can the be the late part if they are the avantguard of Sino-Tibetan in SEA?
"My guess would be O3 in Szechwan and O2 into the coastal provinces".
Uh? And how did it expand from Sichuan? Did the late hunter-gartherers from the highlands suddenly went on rampage and murdered all farmers and then began working and somehow managed to hide all indications of transition?
Burning nail!
"O3 is often considered a marker of Tibeto-Burman expansion".
I crap on that "often-consideration" imagined by some zombie Wikipedian probably years ago, really. O3 as a whole cannot be associated with any modern ethnicity, it's like associating all R1 with Indoeuropeans: we know it's stupid and wrong.
Burning nail!
"But I think we could call the Han movement 'historic'".
Do you mean the occasional colonization and more normal assimilation of peripheral peoples by the nascent Chinese Empire? Yes, that's historical but that is not any major migration.
And someone who is paying (in theory) so much attention to East Asian genetics should know that by now (we have discussed before certainly).
In any case the Han "migration" (assimilation in 90% of the cases) is restricted to South China and the occasional trading outpost in ISEA.
...
"Almost certainly so, but its presence in SE Asia is much more recent".
ReplyDeleteI disagree. We have only a most limited fossil record for all Paleolithic East Asia and nowhere we see a perfectly finished "Mongoloid" or "Sinoid" skull: all look "Australoid" to some extent and North Chinese do too.
On the other hand various "Mongoloid" traits, specially marked cheeks and a tendency to face-flatness and broadness are already present in all them. And I'd be tempted to describe them all (in the North as in the South or in Okinawa) as proto-Mongoloid, just like Crô-Magnon 1 is proto-Caucasoid (with "Mongoloid"/"Alpinoid" features if you wish).
"One minute you're quoting peoples' criticism of Bellwood for proposing too simple a process and here you are proposing just such a simple process".
All I'm saying is that "Mongoloids" (Sinoids) must have been in ISEA long before Neolithic... together with older Negrito groups. These groups do not look neither primarily Austronesian nor primarily Austroasiatic. In fact, I think that Luzon itself was largely Mongoloid before Austronesian arrival and so was the Borneo-Malaysia area before sea level rise.
"Evidence?"
Just look at the previous paragraph: they look like having been there "forever", not to be any offshoot of any mainland population. Add to that Karafet's paper on Y-DNA in the region, that makes most peoples no descedant from Taiwanese Aborigines (proto-Austronesians) nor Indochinese but having their own deep clades.
"Add to that Karafet's paper on Y-DNA in the region, that makes most peoples no descedant from Taiwanese Aborigines (proto-Austronesians) nor Indochinese but having their own deep clades".
ReplyDeleteI've had an idea, but we'll probably need Ebizur's help. It's extremely easy to visualise a convincing scenario for the pattern of origins, and route south, for the three separate Y-hap O strands. Let's see if we can come up with a similarly convincing scenario for their collective origin in the south somewhere.
More than half the Taiwanese Y-haps are O1, specifically O1a. And O1a is more common in southern China than it is in the northern half. The O3 in Taiwan is presumably mostly a product of the Han Chinese movement into the island so we can ignore it for now. Certainly the mainland Chinese language has almost completly replaced the Austronesian languages there. So we might be able to make a reasonable case for O1 having originated in Taiwan, or Fukien, or Kwangtung. But in Taiwan? More than 30,000 years ago? I don't think so. Anyway O1's origin is unlikely to have been anywhere further south than the mainland opposite Taiwan because O1a is very closely associated with Austronesian-speaking people throughout island SE Asia. So almost certainly owes its presence in SE Asia to that expansion.
Let's try O2. It splits into northern and southern clades. The northern clade, O2b, has a recognised tail of three mutations while the southern (O2a) one's tail is just one mutation long. And O2a is associated with many of the non-Han-speaking populations in southern China. O2a is found in Austro-Asiatic and Thai-Kradia speaking people and is common in Vienam, Thailand, Cambodia, and in the western Austronesians of Malaysia and Indonesia. O2a-PK4 even made it way to the west, through the Andhra Pradesh tribals to the Pashtuns. So you may have a case for a southern origin for O2a. But we still have that northern O2b, especially common in northeast China, Korea and Japan, including the Ryukyu Islands. Some O2b does trickle down into Thailand and Vietnam, making a southern origin for the O2 clade possible. But where? O2 could have originated anywhere in the triangle within India, Borneo and Japan. Whether O2 originated in the north or the south it almost certainly reached the other region via the coast. O2 is basically the least common O haplogroup in mainland China.
O3's turn. O3a's diversification certainly seems to have been rapid. Starlike in fact. Unclassified O3a is common in Austronesian-speaking populations, including Polynesians. But surely it's unlikely that some huge Austronesian expansion carried O3a right through SE Asia and into China from Wallacea. So it seems that the basal phylogeny of Y-hap O3a is yet to be resolved. As well as the Austronesian/Polynesian O3a we currently have 7 named clades of O3a: O3a1-M121 in northern China and Cambodia; O3a2-M164 in one person in Cambodia; O3a3-M159 has dissappeared without trace; O3a4-M7 is extremely widespread and common, especially in Tibet and in the Hmong-Mien, and in Malaysia and China; O3a5-M134 in Sino-Tibetan speakers and in the Japanese; and both O3a6-M300 and O3a7-M333 have also dissappeared without trace.
So O3a4 and O3a5 are the main O3 haplogroups to have participated in any original O3a expansion from any single region. But once again it seems we have a north/south split: O3a4 is southern and O3a5 is northern. So we have two O haplogroups that split into northern and southern clades: O2a/O2b and O3a4/O3a5. Coincidently the first clade appears to split near the eastern end of the Chinese Neolithic and the second looks to split near the western end.
Can we arrange the evidence to accomodate a southern origin for Y-hap O?
This is what I see:
ReplyDelete1. O3:
O3 (S. China) -> O3a (S. China) -> O3a3 (Indochina-Sundaland-Philippines) -> O3a3a (S. China) and O3a3b (S. China and Sundaland)
So for O3a there is a flow into SEA proper at the O3a3 stage with backflows at lower levels.
O3 as a whole looks to me original from South China, following Hong Shi 2005.
2. O1:
O1/O1a (Sundaland) -> O1a1 (Sundaland-Taiwan, via mainland probably) and O1a2 (SEA in general, rather rare)
So O1/O1a/O1a1 looks totally original from ISEA with expansion to Taiwan at the O1a1 stage (though this can be argued in the opposite direction but still there would have been migration from ISEA prior to the coalescence of O1a1.
3. O2:
O2a (SEA in general) -> O2a1 (Indochina/S. China)
O2b (North and Coastal East Asia/SEA) -> O2b1 (Coastal East Asia/SEA)
You raise the question of the origin of O2 as a whole but we can by the moment just draw a generic convenient centroid in SE China.
So we have three centroids at, say, Guangzhou (O2), Hunan (O3) and Jakarta (O1). The overall centroid could well be near the Mekong delta, I'd say (Cambodia or Cochinchina). However as 2/3 clades seem centered in South China, you might argue for a slightly more northerly origin but in Indochina in any case, I'd think.
Thanks for you suggestions.
ReplyDeleteIf Ebizur is reading this I'm sure he will be able to supply the necessary information.
"O3a3a (S. China) and O3a3b (S. China and Sundaland)".
And O3a3c in the Sino-Tibetan and Japanese people. But that scenarion puts the very strong representaion of O3a through the whole of China and even in Japan as a recent migration north from SE Asia, for which there is no evidence whatsoever.
"So O1/O1a/O1a1 looks totally original from ISEA with expansion to Taiwan at the O1a1 stage (though this can be argued in the opposite direction but still there would have been migration from ISEA prior to the coalescence of O1a1".
Again we have problems. O1a-M119 is found in the Kradai speakers as well as the island SE Asia population. So once more we need a recent migation north for which there is no evidence.
"You raise the question of the origin of O2 as a whole but we can by the moment just draw a generic convenient centroid in SE China".
But to accomodate the northern O2b we have to postulate a migration north again.
Why "recent"? For me all that is Paleolithic stuff.
ReplyDelete"Again we have problems. O1a-M119 is found in the Kradai speakers as well as the island SE Asia population. So once more we need a recent migation north for which there is no evidence".
I did not have the source for that. I used Hong Shi'05 and Karafet'10 for my reconstruction.
Anyhow, it'd be interesting to know more details about the structure of O1 before we issue judgment. Whatever the case O1a in ISEA does not look at all Austronesian, though admittedly it might have originated in mainland SEA (Indochina or South China).
"Anyhow, it'd be interesting to know more details about the structure of O1 before we issue judgment".
ReplyDeleteDo you know how to get in touch with Ebizur? He could probably help us. I have an idea he's something to do with the quetzacoatl blog.
An important detail is that while MNOPS as whole looks totally as original from Sundaland (per the geography of its descendants), O looks somewhat more northernly, mainlander.
ReplyDeleteSo when O1 and O3 arrived to West Indonesia, they must have done on an older layer, preserved best among Negritos.
But I do not think this migration can be considered Neolithic. The exact dates should be assessed on archaeological grounds (when these finally are better known than today) but at most I'd argue for a late Paleolithic (post-Glacial Maximum) date.
A post-LGM date could be interesting because, if I'm correct, the main technoculture of the area, the Hoabinhian, is of at least those dates and seems to originate in Indochina.
No, I don't think I have his email. Sorry. Surely his input would be valuable indeed.
ReplyDelete