While this conclusion was something more or less reachable with previous data (see HERE for example), a new study adds some fine detail for us to reconstruct the paleohistory of this major Eurasian lineage.
Peter A. Underhill et al., The phylogenetic and geographic structure of Y-chromosome haplogroup R1a. EJHG 2014. Pay per view → LINK [doi:10.1038/ejhg.2014.50]
Important: supplemental materials are freely available.
Abstract
R1a-M420 is one of the most widely spread Y-chromosome haplogroups; however, its substructure within Europe and Asia has remained poorly characterized. Using a panel of 16 244 male subjects from 126 populations sampled across Eurasia, we identified 2923 R1a-M420 Y-chromosomes and analyzed them to a highly granular phylogeographic resolution. Whole Y-chromosome sequence analysis of eight R1a and five R1b individuals suggests a divergence time of ~25 000 (95% CI: 21 300–29 000) years ago and a coalescence time within R1a-M417 of ~5800 (95% CI: 4800–6800) years. The spatial frequency distributions of R1a sub-haplogroups conclusively indicate two major groups, one found primarily in Europe and the other confined to Central and South Asia. Beyond the major European versus Asian dichotomy, we describe several younger sub-haplogroups. Based on spatial distributions and diversity patterns within the R1a-M420 clade, particularly rare basal branches detected primarily within Iran and eastern Turkey, we conclude that the initial episodes of haplogroup R1a diversification likely occurred in the vicinity of present-day Iran.
This case, as well as many others, including that of its close relatives R1b and Q, illustrate why frequency is not the same as origin, which can only be inferred (if at all) by studying the hierarchical diversity of the lineage. These three lineages for example, must have spread from West Asia but they are relatively less important in numbers in that region today, overshadowed by other lineages, notably J. Instead their derived branches had major impacts in other regions (Europe, South and Central Asia, Siberia and America).
Frequencies of the main lineages
There are two main sub-lineages of R1a, which according to the current ISOGG tree version (maybe to be refitted after this study?) are known as R1a1a1b2 (Z93) and R1a1a1b1a (Z282). The first one is essentially Asian (with greatest frequencies in South and Central Asia, where it includes >98% of all R1a individuals) wile the latter is almost exclusively European (notably Eastern European but with a distinct branch in Scandinavia, encompassing together >96% of R1a individuals in Europe).
These maps give us a quite decent glimpse of the main scatter patterns of R1a but alone they can't inform us of its origins. For that we have to look at the detailed tree and the relationship of its samples with geography.
Origins and distribution of R1a
As mentioned above, the authors conclude that R1a and R1a1 must come from Iran, where the greatest basal diversity is:
To infer the geographic origin of hg R1a-M420, we identified populations harboring at least one of the two most basal haplogroups and possessing high haplogroup diversity. Among the 120 populations with sample sizes of at least 50 individuals and with at least 10% occurrence of R1a, just 6 met these criteria, and 5 of these 6 populations reside in modern-day Iran. Haplogroup diversities among the six populations ranged from 0.78 to 0.86 (Supplementary Table 4). Of the 24 R1a-M420*(xSRY10831.2) chromosomes in our data set, 18 were sampled in Iran and 3 were from eastern Turkey. Similarly, five of the six observed R1a1-SRY10831.2*(xM417/Page7) chromosomes were also from Iran, with the sixth occurring in a Kabardin individual from the Caucasus. Owing to the prevalence of basal lineages and the high levels of haplogroup diversities in the region, we find a compelling case for the Middle East, possibly near present-day Iran, as the geographic origin of hg R1a.
Between these top tier nodes (R1a and R1a1) and the two most common sublineages described above, this study only found one paragroup represented: R1a1a1* (M417). This should be an important step in the analysis but the researchers prefer to remain silent on it. Why? I guess that the reason is that it is complicated to analyze and reach to sound conclusions.
I spent some time today looking at the haplotypes of this paragroup mentioned in the study and I could not reach a conclusion either: the majority of the sequences are from Europe and all them (excepting a highly derived Norwegian line and including a low derived Iranian one) seem to derive from a North German haplotype. I call this group "branch A".
However there is at least one West Asian sequence (from Turkey) which seems independent ("branch B"), while an Indian and the already mentioned Norwegian sequence could derive from either one. So my impression is that there is an specifically North European "branch A" but also some other stuff with West Asian centrality ("branch B") within this key paragroup.
Guess that I could say a lot more about not being able to say much more on this key intermediate step but, synthetically there are two options among which I can't decide:
- Branch A went back to West Asia from where it spread again to Eastern Europe and Central South Asia.
- Branch B is actually at the origin of the two derived and highly spread subhaplogroups.
Whatever the case I understand that there are good reasons to think that these spread first from West Asia, at the very least Z93 and very likely also Z282.
R1a1a1b2 (Z93)
There is nothing European in this lineage: only some lesser terminal branches at the Southern Urals, roughly where the Kurgan phenomenon began some 6000 years ago.
This detail is indeed remarkable because, if, as often argued, R1a or some of its subclades spread from there, we should expect at least some basal diversity being retained. Instead all we see are some highly derived branches. So the main conclusion must be that the expansion of R1a does not seem related to the Kurgan phenomenon, except maybe in some secondary instances.
As mentioned before, this lineage is Central and South Asian and comprises the vast majority of R1a in those two regions.
The detailed haplotype network can be seen in Supp. Info fig. 2.
In essence we can say that:
- Z93* has three apparent distinct branches stemming from West Asia (incl. Caucasus) and another one from South Asia/Altai (1).
- Z95* has two apparent distinct branches:
- A small one with presence in West Asia and Southern Europe
- Another one (pre-M780?) stemming from South or West Asia
- M780 has clear origins in South Asia (incl. most Roma lineages)
- Z2125 also appears to originate in South Asia, even if it has a greater spread outside it, notably to Central Asia
- M580 and M582 appear related and surely originated in West Asia
Weighting them:
- Z95:
- West Asia: 2
- South Asia: 2
- West/South Asia: 1
Therefore the origin of Z95 should be though as West-South Asian but undecided between either region. Say Afghanistan for example.
- Z93:
- West Asia: 3
- West/South Asia: 1 (Z95)
- South Asia/Altai: 1
In this case I would say that West Asia is almost certainly the origin, although tending to Central/South Asia. For example: Iran again.
So, regardless of whether the previous stage (M417) represents a stay in West Asia or a back-migration from Europe into West Asia, West Asia is clearly at the origin of Z93. It does not represent any Kurgan migration but an Asian phenomenon with origins towards the West (around Iran).
R1a1a1b1a (Z282)
On first sight this European sublineage seemed quite simpler: it is obvious that the bulk of it spread from Eastern Europe. However, when we look at the haplotype network, we cannot confirm this pattern for the Norwegian or Scandinavian haplogroup Z284, which is only linked to the rest via some South European and West Asian samples.
So my conclusion must be that Z282 experienced a main expansion from Eastern Europe but only into Eastern and Central Europe and that the Scandinavian variant almost certainly represents another flow within this haplogroup, with the knot being in West Asia.
Anyhow the main East and Central European expansion seems true. For some reason it is not centered in any obvious prehistorical locality, as could be the Volga or maybe Ukraine, but instead its center is further North around Smolensk.
Overall reconstruction of the spread of R1a
With all the previous analysis I made this map, which also shows in discrete gray color the general pattern of expansion of haplogroup R:
We have an expansion of R into South Asia and Western Eurasia (incl. Central Asia) and even into parts of Africa (R1b-V88) from apparent South Asian (R, R1 and R2) and West Asian (R1a, R1b) origins. Related lineages Q and P* could also be integrated into this pattern of expansion but I did not want to overload the map with too many details.
There is some uncertainty regarding the North European branches of R1a but otherwise the pattern seems quite clear.
On these North European branches, I must say that they remind me of other odd lineages with similar geography: R1b-U106, I1-M253 and I2a2-M223. With the likely exception of R1b-U106 neither appears to have experienced any significant re-expansion since their arrival to that corner of the World, however they do seem to survive pretty well in it.
Time frame?
Finally we seem to be entering the age of full Y chromosome sequencing and a more serious molecular clock based on it. As I have explained on other occasions (for example), the human Y chromosome is large enough to experience mutations almost every single generation, what should provide a decent molecular clock, unlike the very rough approximations used in the past.
However the issue of correct calibration remains open. As you surely know the academy is slow to incorporate the most recent evidence, especially from fields distinct to their specialty. Hence I do not expect them to calibrate based on the obvious fact that age(CF) or at least age(F)=100,000 years. They are probably still stuck in old concepts of a "recent" out-of-Africa migration c. 60 or at most 80 Ka ago, as well as the usual Pan-Homo spilt under-estimates
I must reckon in any case that I had not enough time to study this matter in depth yet, so the previous observation is rather my idea of what to expect.
In any case in this study the authors resorted to full Y chromosome to calculate their age estimates and I applaud them for doing so. As apparent in fig. 5, all R1 derived sequences have approximately the same number of accumulated SNPs, what in principle allows for a perfected molecular clock, assuming it is well calibrated.
Their estimate is as follows:
A consensus has not yet been reached on the rate at which Y-chromosome SNPs accumulate within this 9.99Mb sequence. Recent estimates include one SNP per: ~100 years,⁵⁸ 122 years,⁴ 151 years⁵ (deep sequencing reanalysis rate), and 162 years.⁵⁹ Using a rate of one SNP per 122 years, and based on an average branch length of 206 SNPs from the common ancestor of the 13 sequences, we estimate the bifurcation of R1 into R1a and R1b to have occurred ~25,100 ago (95% CI: 21,300–29,000). Using the 8 R1a lineages, with an average length of 48 SNPs accumulated since the common ancestor, we estimate the splintering of R1a-M417 to have occurred rather recently, B5800 years ago (95% CI: 4800–6800). The slowest mutation rate estimate would inflate these time estimates by one third, and the fastest would deflate them by 17%.
The references correspond to (4) Poznick 2013, (5) Francalacci 2013, (58) Xue 2009 and (59) Méndez 2013. This last is the Anzick study, of which at the very least we can say that they had a real calibration point in the ancient Amerindian DNA. It is also the one which provides the longest mutation rate.
Considering that Xue 2009 is "old" (for this avant-guard aspect of this pretty young science), I find their choice of the Poznick rate quite a bit conservative. The Francalacci rate is the intermediate one of the three "recent" papers referenced and it is also quite close to the calibrated Méndez rate.
Personally I would choose the later without a second thought. As long as CF ends up being younger than 100 Ka, it is positively too conservative anyhow.
Using the Méndez (Anzick-calibrated) rate of 162 years per SNP, I get the following corrected estimates:
- R1a/R1b split (R1 node): 33,000 years ago (CI: 26.0-42.5 Ka)
- R1a-M417 node: 7,700 years ago (CI: 6.4-9.0 Ka)
These seem fair enough to me, judging on the fact that the core R1a expansion seems to originate in West Asia (at the very least for the South/Central Asian branch), what fits much better with a Neolithic frame than with the Kurgan one.
It also fits better with my previous estimates after due re-calibration of Terry D. Robb's full sequence Y-DNA tree, although my estimates are even older, especially after a second recalibration to adjust to the recent discovery of widespread H. sapiens evidence in South and East Asia c. 100 Ka ago.
In my understanding the R1 node is actually c. 48 Ka old (R1b: c. 34 Ka.), what, apportioning, yields a date of c. 11.2 Ka for the R1a-M-417 node.
Update (Mar 31):best possible molecular clock estimates for R1:
Follows fig. 5 of Underhill et al. 2014, annotated by me in red and purple colors:
If I'm correct, then the expansion of R1b in Europe still corresponds in rough terms to the Magdalenian period or, more generally, the late Upper Paleolithic. This does not mean that it remained that way forever (it may well have been reshuffled later on: in the Epipaleolithic, Neolithic and Chalcolithic) but it seems to be the time-frame of its main expansion when the main lineages got established, whatever happened to them later on.
I know well that so far ancient DNA for this lineage remains to be found and that the dominant haplogroup among known Epipaleolithic hunter-gatherers was (for all we know) I2a. However this is what the refined full Y chromosome sequence molecular clock, properly calibrated according to the archaeological evidence for the settling of Asia by H. sapiens, has to say. If you wish to dismiss this and use another estimate instead, that's always up to you. I just hope that you know what you're doing.
Anyhow, if I am correct, then the expansion of R1a is neither Chalcolithic nor Neolithic but clearly Epipaleolithic. Does it make any sense? I can't say for sure because this period is not so well understood. Whatever the case, is it possible to integrate the key pre-Neolithic Zarzian culture of the Zagros (map) in this scheme of things? What about all the other question marks that fill the gaps of our mediocre knowledge of the Mesolithic of West Asia? Or is it the Balcanic Epigravettian to be blamed instead? Or both?
I really can't say with any certainty at this stage. But I am intrigued indeed.
Update (Mar 31): frequency pie charts of Underhill's data available at Kurdish DNA.
Update (Aug 2015): I must update the frequencies of the various upstream paragroups, in agreement with table S4, because I may have missed some details initially. However the overall tendency is the same.
Update (Mar 31):best possible molecular clock estimates for R1:
Follows fig. 5 of Underhill et al. 2014, annotated by me in red and purple colors:
- Red: age estimates calibrated for age(CF)=~100Ka, which is what modern archaeological evidence overwhelmingly supports for the second phase of the migration Out of Africa (or rather out of Arabia into India and East Asia). Dots drawn to help identify the estimated nodes.
- Purple: general references of European (plus) prehistorical cultures or periods for the key ages estimated.
If I'm correct, then the expansion of R1b in Europe still corresponds in rough terms to the Magdalenian period or, more generally, the late Upper Paleolithic. This does not mean that it remained that way forever (it may well have been reshuffled later on: in the Epipaleolithic, Neolithic and Chalcolithic) but it seems to be the time-frame of its main expansion when the main lineages got established, whatever happened to them later on.
I know well that so far ancient DNA for this lineage remains to be found and that the dominant haplogroup among known Epipaleolithic hunter-gatherers was (for all we know) I2a. However this is what the refined full Y chromosome sequence molecular clock, properly calibrated according to the archaeological evidence for the settling of Asia by H. sapiens, has to say. If you wish to dismiss this and use another estimate instead, that's always up to you. I just hope that you know what you're doing.
Anyhow, if I am correct, then the expansion of R1a is neither Chalcolithic nor Neolithic but clearly Epipaleolithic. Does it make any sense? I can't say for sure because this period is not so well understood. Whatever the case, is it possible to integrate the key pre-Neolithic Zarzian culture of the Zagros (map) in this scheme of things? What about all the other question marks that fill the gaps of our mediocre knowledge of the Mesolithic of West Asia? Or is it the Balcanic Epigravettian to be blamed instead? Or both?
I really can't say with any certainty at this stage. But I am intrigued indeed.
Update (Mar 31): frequency pie charts of Underhill's data available at Kurdish DNA.
Update (Aug 2015): I must update the frequencies of the various upstream paragroups, in agreement with table S4, because I may have missed some details initially. However the overall tendency is the same.
- R1a* (M420): Italy (1), Turkey East (1), Turkey Cappadocia (2), UAE (1), Oman (1), Iran (set 2) (2), Iran NE (1), Iran South (5), Iran North (5), Azeris-Iran (5).
- R1a1* (SRY10831.2): Iran (set 2) (1), Iran NE (1), Iran South (2), Iran North (1), Kabardin (1). In addition it has more recently been found in two Epipaleolithic Eastern Europeans (EHG), from Karelia (Haak 2015) and Smolenskaya Oblast (Chekunova 2014).
- Ra1a1a1* (M417): Ireland (1), Netherlands (3), Norway (1), South Sweden (1), Germany (1), Estonia (1), Hungary (1), Turkey East (Kurds) (1), Iran (set 3) (1), India South (1).


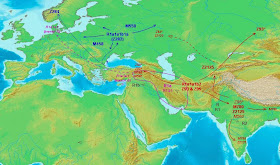

R1a mostly spread from here during the Copper Age, between the modern hotspots of Z282 and Z93, along with ANE.
ReplyDeletehttp://i129.photobucket.com/albums/p217/dpwes/East_Euro_K15.png~original
That's the East Euro component from the Eurogenes K15. MA-1 carries 34.45% of it.
One of the problems with Underhill et al. 2014 is that the phylogeography of European R1a is a mess, with, for instance, Z280 being shown to be above Z282, M458 and Z284.
Another problem is that the M420* samples from the Near East appear to belong to single young subclade, so they're not evidence of an Iranian origin of R1a, especially since M420* is also found across Europe, except it wasn't reported from there in this study.
So your map doesn't make any sense. At some point you'll realise that when you start thinking of R1a as an ANE marker.
We are talking Y-DNA and you want to "prove" something about it with autosomal DNA which seems essentially Uralic? Sorry but I don't get your point.
Delete"One of the problems with Underhill et al. 2014 is that the phylogeography of European R1a is a mess, with, for instance, Z280 being shown to be above Z282, M458 and Z284".
Is it a problem or a "solution"? I understand that they did not find any differential sequences within those samples but maybe I misunderstood something, did I?
"Another problem is that the M420* samples from the Near East appear to belong to single young subclade, so they're not evidence of an Iranian origin of R1a, especially since M420* is also found across Europe, except it wasn't reported from there in this study".
M420* (R1a1*) has only been reported AFAIK in Greece (almost Asia) and Scandinavia (the cul-de-sac where all oddballs end piling up). That's very different from "across Europe". Obviously its frequencies are tiny enough to be irrelevant and lack enough STR diversity to matter. You can't build up a counter-theory on mere erratics.
The authors tested STR diversity, so the idea you want to push of "the M420* samples from the Near East appear to belong to single young subclade" doesn't seems to stand to scrutiny. Of course, if you can demonstrate it properly, I will be willing to read your article on the matter but, sincerely, I doubt you can.
My impression is that you are just pushing your preconceptions (ideology?) by sowing confusion. That's also the impression I got in the previous discussion we had on the matter.
The last thing you said in your usual cryptic style was: "Don't worry, you'll get it eventually when more stuff comes out. No point arguing about it now".
Well, here there is more stuff, and as far as I can discern it proves me right actually.
"At some point you'll realise that when you start thinking of R1a as an ANE marker".
That doesn't make any sense: Iberians are like 30% more ANE than Basques but they don't have almost any R1a. Estonians are more ANE than Poles but they have less R1a, etc. Basques more than double Sardinians in ANE affinity but we do not have any more R1a than they do (if anything the opposite is true).
You should try to segregate haploid genetics from autosomal genetics. The latter may represent "recent" flows and endogamous homogenizations (not always easy to discern anyhow) but the former often informs us of ancient patterns which are usually blurried in the autosomal data and the phylogeny, if properly done, is "God's word", so to say - nothing to misinterpret in it.
Also the autosomal data can be very imprecise, confuse and contradictory; in most cases I do not see any sort of cross-validation checks that justify the choice of K-level, in other cases I see how small endogamous populations cope the clusters: almost one for each, while large more exogamous ones remain undifferentiated. It is a data point but you can't read too much in it, especially not if you are careless about cross-validation and do not test your hypothesis with formal f3 tests, something seldom done. After all it is just an algorithm designed to simplify massive amounts of highly complex data into a simplified visual graph, and every simplification departs from reality. The map is not the territory.
The east to west expansions of R1a, ANE and Indo-European languages into Europe are all dated to the Copper Age via different but increasingly accurate means. They obviously all came with the same people, who also spread to India at the same time.
DeleteOn the other hand, the set of events you're arguing for here never happened, and you'll realize that very soon.
Obviously there seems to be some sort of apparent contradiction with the increased levels of ANE affinity, which can be attributed to IE spread and the lack of anything similar in the haploid side. Even if we'd accept R1a as such marker, it'd be unable to explain the variable ANE affinity levels on its own.
Delete"... and you'll realize that very soon".
I would realize (assuming there's anything to realize) if you or someone would be able to demonstrate. So far I only or mostly see empty "prophecies".
Davidski likes throwing out stupid objections, just to stall the demise of a Steppe fantasy - one which sound discourse has long defeated.
DeleteMaju, I would be very happy if you could check if they included Saami R1a in the study? Is it possible to know if Saami R1a belongs to Z282 or Z284? It seems that the Finnish R1a is split between Z282, Z284, M458 and M558.
ReplyDeleteDo they give an age estimation for M458 or for M558?
I would suppose that the Scandinavian Z284 does not have anything to do with the steppe IE languages but spread to the north during the Neolithic. Instead, M558 could be related to Steppe IE phenomenon and there are certain correspondences between the spread of M558 and the Corded Ware culture. Am I right that M458 has been proposed to be the Slavic marker (but not the only one). On the basis of the frequency map, it could be true.
Maju, you propose that R1a spread to Europe through Turkey. If R1a arose in Iran or in Afghanistan, I would prefer a route to Europe through Daghestan or even East of the Caspian Sea.
For Corded Ware maps:
http://en.wikipedia.org/wiki/Corded_Ware_culture
https://www.google.fi/search?hl=fi&site=imghp&tbm=isch&source=hp&biw=1165&bih=701&q=%22corded+ware%22&oq=%22corded+ware%22&gs_l=img.3...0.0.1.180761.0.0.0.0.0.0.0.0..0.0....0...1ac..38.img..6.7.730.wTxaK95RHh8
I don't see any Saami sample in the supplemental info (anyhow freely available). I can't see any Finnish sample either. The dots in the maps should represent samples from table S4 but maybe they forgot to parse them?
DeleteThere are a number of other Finnic samples however:
Estonians: M417*: 0.4% (IMO derived from the North German haplotype and close to the South Dutch one), Z282*: 6.4%, M458: 5.1%, M558:19.1%, Z284: 0.4%, Z93*: 1.7%, Z95 and downstream: 0.0%
Vepsas: M417*: 0.0%, Z282*:5.1%, M458: 15.4%, M558: 15.4%, Z284: 0.0%, Z93 and downstream: 0.0%
Karelians: M417*: 0.0%, Z282*: 16.4%, M458: 13.6%, M558: 10.0%, Z284: 0.0% Z93*: 0.0%, Z95*: 0.7%, downstream of Z95: 0.0%.
"Do they give an age estimation for M458 or for M558?"
Not explicitly but the nodes for Z282, Z93 and M780 seem not far from the the main M417 one (seems a double star-like spread), so they should be only slightly more recent.
"I would suppose that the Scandinavian Z284 does not have anything to do with the steppe IE languages but spread to the north during the Neolithic".
My impression from the haplotype structure suggests a separate flow from West Asia and/or Southern Europe but the info is thin enough to allow for some doubt.
"Instead, M558 could be related to Steppe IE phenomenon".
I can think of both M558 and M458, as well as the bulk of Z282*, representing a single expansion from around Smolensk (?) These are the best fit with the Kurgan phenomenon but I feel that, unless new data comes around, the more reasonable estimate dates (in my opinion) suggest an older expansion than Chalcolithic and maybe even older than Neolithic.
I admit however that I do have a problem explaining this estimate chronology for Eastern European R1a. But maybe we are missing something important in the archaeological record.
"Maju, you propose that R1a spread to Europe through Turkey. If R1a arose in Iran or in Afghanistan, I would prefer a route to Europe through Daghestan or even East of the Caspian Sea".
I reckon that I did not pay much attention to the Caucasus, largely because the inclusion of Armenia in it or the lack of distinction between N/S Caucasus in the categories, made it difficult to discern what is West Asian and what specifically Caucasian without looking at the fine detail, so I tended to lump both regions in the same simplified one.
But I did not identify any Caucasus labeled node in the Z282 haplotype network in any relevant position, so I drew the dotted arrow via the Balcans ("South Europe") instead.
As for upstream stuff I did not identify any possible Caucasus-specific route either, nor Central Asian one in either case, except for the occasional highly derived end-of-branch haplotype.
So for me there is little question that the main arch of Eurasian distribution of R1a in general and its subclades as well goes through the Middle Eastern "highlands", between Turkey and Iran, linking to South Asia via AfPak and to Europe via the Balcans.
So, you think Z93* in the Altai comes from south Asia/south central Asia?
ReplyDeleteI think it would make more sense to link this R1a subclade to the arrival of the Europoid population appearing during chalcolithic (origin of Afanasevo) in south Siberia, a population clearly coming from south Russia/south-east of the Urals, from the eastern part of the Yamnaya peoples (common morphology, light pigmentation, _west eurasian_ (with modern european matches) female lineages, kurgans, early Yamnaya potteries and cultual objects and even axes, copper metallurgy, pastoralism (linked w/ modern cattle DNA in Mongolia and beyond w/ a sizeable European component), typical dental characteristics, and so on).
If this R1a-Z93* arrived from south Asia, how comes it is largely associated with typical _WESTERN_ female lineages (sometimes with modern matches as far as Iceland, in the case of a mtDNA H of bronze age Tarim).
How do you explain it?
If you don't associate it with the beginning of Afanasevo what are you associate it with?
Are you envisionning some kind of population replacement? Are you thinking of a wiping out when Afanasevo became part of the Andronovo horizon around 1700 BCE? It doesn't change much anyway, Y-DNA-wise as the source of andronovo is also ultimately the kurgan culture of Russia.
This europid Z93* in the Altai, associated with west eurasian female lineages (and not south Asian ones), seems to corroborate the Kurgan theory more than anything, since it links Z93's ancestor with south Russia's ancient eastern Kurgan cultures.
"So, you think Z93* in the Altai comes from south Asia/south central Asia?"
DeleteOr West Asia maybe. As I said above, "say Afghanistan" but could also be Iran or Pakistan or whatever in that knot area. The lack of known direct precursors anyhow (M417*) is a bit problematic because, looking at the haplotypes (not in full depth, except for M417* itself) the link may seem to be rather to the West, even in Europe. Judging from the modal Z93 haplotype 15-12-13-17-25-11-10-10:
→ N. German: 1 STR step away)
→ Turk ("branch B"): 2 steps
→ North Dutch and Iranian: 2 steps but apparently via the N. German haplotype (i.e. "branch A")
This part is the most difficult one and the authors do not address it at all.
But whatever the case the Z93 haplotype network points to very basal South/West Asian branches dominating the structure, so it seems reasonable that the origin is towards the South.
"I think it would make more sense to link this R1a subclade to the arrival of the Europoid population appearing during chalcolithic (origin of Afanasevo) in south Siberia"...
It'd be easier to explain maybe but I do not see how: the modal Z93* haplotype is quite divergent from the M417* ones, be them European or Asian and mostly Z93* seems to be a couple of yet undescribed sub-haplogroups (plus a few southern smaller branches maybe).
Please look at the haplotype structure: it is important.
If the older molecular clock estimates, not my own but the ones derived from Méndez' or Francalacci's studies, which have been too lightly sidelined, are correct, then the time-frame would be approximately Neolithic, what is ideal to explain an expansion from West Asia, as it seems to be the case. In the case of South and Central Asia I see a very good fit for this Neolithic model. Also prolific farmers would be much more likely to cause a demic impact against the hunter-gatherer precursors than a bunch of Bronze Age raiders versus one of the greatest civilizations of that time.
As for pigmentation, the first known modern genetics (many doubts about the resulting phenotype) are from early european farmers, so it's quite parsimonious that they also brought similar genetics to South Asia, although there selection rather acted against than in favor of lighter skin colors.
"If you don't associate it with the beginning of Afanasevo what are you associate it with?"
Isn't Central Asia "Western" (not necessarily meaning "European") since the very beginnings of the Upper Paleolithic? I don't need any particular explanation for Altai or other Central Asia aligning with Western genetics all the time before the Turkic migrations of the Iron Age: it's what I would expect considering its cultural links (Aurignacoid, Gravettian, Western Neolithic, etc.)
"Are you envisionning some kind of population replacement?"
Seems likely in a Meso-Neolithic time-frame wherever R1a is important in frequency.
"Are you thinking of a wiping out when Afanasevo became part of the Andronovo horizon around 1700 BCE?"
I see no genetic hints of these cultural phenomenons significantly affecting the genetics of those areas: Central Asia and Eastern Europe are clearly two different things R1a-wise.
"It doesn't change much anyway, Y-DNA-wise as the source of andronovo is also ultimately the kurgan culture of Russia".
One thing is culture, ethnicity and language and another one genetics. It's perfectly possible to radically alter the ethnicity of a population with a very minor genetic impact, via elite domination (plus some time and some luck maybe). Jamaicans or Haitians are Indoeuropeans but genetically they have almost no relation with Europe, Spaniards or French are "Romans" but the genetic impact from Italy in those areas is tiny at most.
"This europid Z93* in the Altai"...
It is not "Europoid" in any way I can discern: just look at the haplotypes, for Chaos shake!
@Vooruit, if Tarim had Z93 on male line and European mtDNA, maybe they were West/Central Asian migrants who favored European wives (like the Ottoman sultans not so long ago). Motif isn't so unusual (some modern pops have it and everyone gets confused by trying to awkwardly classify them as Euro or Mideastern when they are typical of both and neither).
DeleteEven their ethnonym Tokhri sounds Anatolian. Like Taurus Mountains or Taurica. Maybe some West Asians got squeezed out to Central Asia and stayed there.
We cannot take apart European from West/Central Asia mtDNA so easily, much like we could not discern European and South/Central Asian yDNA R1a until recently. Unlike Y-DNA, mtDNA cannot be near-infinitely split into new subclades.
DeleteSo no need to be making up complicated fantasy harem tales that are not likely to be real.
First let me say as someone who hasn't read the paper , this is an informative blog-post, except for:
ReplyDelete“Hence I do not expect them to calibrate based on the obvious fact that age(CF) or at least age(F)=100,000 years.”
This is just unsupported nonsense.
“As long as CF ends up being younger than 100 Ka, it is positively too conservative anyhow. “
I don't think this type of unwarranted confidence is helpful at all Maju, it sounds as if ancient YDNA from greater than 100 KYA has been found with the CF-P143 mutation, has there? Am I missing something?
Don't forget that mtDNA from ancient sites has more or less vindicated the orthodox views on molecular 'clockology' as you like to refer to it, we are just waiting for YDNA to do the same.
"This is just unsupported nonsense".
ReplyDeleteAnd that is an unsupported and very gratuitous disqualification.
There's a lot of recent archaeological and paleontological evidence piling up that clearly point to an arrival of H. sapiens to South and East Asia c. 100 Ka ago. Link in main entry.
Also it makes good sense if we consider the "pump" model for the OoA migration: when conditions were favorable in the Abbassia Pluvial, people moved to the "deserts" (then much more productive) of Sahara and Arabia. Lots of archaeological evidence confirm it since c. 125 Ka ago. When the Pluvial was declining, some of them may have been pushed in search of new opportunities, reaching to Asia East of the Arabian Sea and rapidly expanding in that area.
"Am I missing something?"
Yes: you are totally missing the archaeological evidence, which is the only one informing us about the Out-of-Africa migration time frame in fact.
"And that is an unsupported and very gratuitous disqualification."
DeleteIt just means that your speculation with respect to 100 KYA age for either Node CF or F makes very little sense with respect to almost all pertinent studies done to date, hence it is unsupported, and your assertion that it is 'obvious' is devoid of any sense, that is what I meant.
Any way, it looks like older nodes, like R1, give TMRCA estimates using SNPs that are closer to using the zhivotovsky rates for STRs, while the younger nodes, like R1a, gives estimates closer to using the pedigree rates.
5736 R1 haplotypes from FTDNA
Underhill (2014) R1a haplotypes
Y TMRCA calculator
All that is irrelevant: only archaeology and paleoanthropology can provide valid calibration points. All the rest is circular logic, a trap in which scholasticism falls a lot.
DeleteFrom genetics we can only know the proportions of the branches but the "units" that measure them are abstract and in order to translate them to realistic time units we need realistic calibration points, which are seldom genetic (at the very least it must be ancient DNA).
“All that is irrelevant: “
DeleteIt is relevant because it steers us onto the correct direction of which predictive Y TMRCA calculating model to use, and since the primary variable that all these models hinge upon is the mutation rate, it is logical to study the impact of different mutation rates on different models, all ancient DNA is going to do is corroborate (or not) one of these mutation rates, the models will still be used to compute TMRCAs in the future, hopefully with better accuracy due to refined mutation rates.
So, even-though I have very limited knowledge in the field of European YDNA, this is my primary interest;
In this study , which used the SNP counting model
R1 is predicted to be 20.6 - 33.4 KYA (based on the 4 mutation rates that you show)
Publicly available YSTR data from ftdna (see my previous post) however shows for R1,
using Zhivotovsky (effective rates): 26.5 – 38.1 KYA
using Pedigree rates (from various sources): 10.2 – 22.0 KYA
Next when we look at the younger R1a,
The SNP counting model shows:
R1a : 4.8 – 7.8 KYA
While YSTR data from ftdna for R1a shows:
using Zhiv: 13.4 KYA
using Pedigree: 4.3 – 7.6 KYA
The YSTR data for R1a from this study (Underhill 2014) shows relatively similar results to ftdna:
using Zhiv: 12.5 KYA
using Pedigree: 3.2 – 4.9 KYA
So , we can easily see that while the Zhiv based YSTR estimates show closer estimates to the results of the SNP counting model for R1, this is not the case for R1a, in fact, the pedigree rates are a better predictor of the results of the SNP based model. The question off-course is why is this?
You know well (or should know) that I have always been extremely critical of Y-STR "molecular clock", which I considered a heap of junk: a self-complacent pseudo-method without any support whatsoever and with exactly zero predictive power.
DeleteFull sequence Y-chromosome is completely different: it is the first approximation to time estimates in which I can place any hope because after all, there's not much time for demographically-led rearrangement between mutation and mutation.
"The question off-course is why is this?"
Not sure: probably because microsatellites by definition could only be a mediocre approximation (too few markers: this is megas of info instead!): much ado about nothing,
And that was what I always insisted on: all you think you know about Y-DNA age estimates is almost certainly wrong. For me that's cool and expected, for you I guess it can be frustrating at first - but you should get used after some time for reprocessing and maybe even enjoy it even more than I ever could.
Whatever the case, there is a problem shared by both methods: proper calibration. If they don't pay attention to the OoA archaeology or the early hominins data for the Pan-Homo split, I can only hope that they at least pay some attention to ancient DNA sequences.
“For me that's cool and expected, for you I guess it can be frustrating”
DeleteOn the contrary , I keep a wide perspective on TMRCA estimates, that is why I compute estimates based on both the pedigree and effective rates and don't have a bias towards anyone of them. You on the other hand are fixed in your ways, because when the numbers don't agree with you, you just make up mutation rates on the fly, for instance in this current blog post you propose R1 to be 48 KYA, which would convert to one SNP per 233 years, which is 44% slower than the slowest mutation rate , and where did you come up with this mutation rate, absolutely no where, you just came up with a number that fits your own little archaeological views, even though that may be acceptable to you it certainly is not how it works.
“I can only hope that they at least pay some attention to ancient DNA sequences.”
Yes, me too. But the other question is will you pay attention to the results, if they do include ancient DNA sequences. For instance, I still clearly recall that when Fu (2013) came out with ancient DNA calibrated MRCA estimates for mtDNA, largely congruent with previous estimates, you were still amazingly defiant of the results.
... "where did you come up with this mutation rate, absolutely no where, you just came up with a number that fits your own little archaeological views"...
DeleteI came up with a number that fits the archaeological FACTS.
How can you be so dismissive of archaeological data. That can only be described as blind arrogance, Ethio.
"... it certainly is not how it works".
It is exactly how it does work in fact.
A decade ago or two, the archaeological paradigm suggested a migration out of Africa c. 70-50 Ka and the population geneticists calibrated their primitive molecular clocks according to those ideas, as well as to similarly obsolete ideas about a very recent Pan-Homo split some 5-7 Ma ago.
Since then archaeology and paleontology has advanced a lot but population genetics' molecular clock-o-logy has remained scholastically fixated in their own intra-disciplinary references, references to references and references to references to references. It's a completely vicious circle of the worst scholasticism possible.
It may be relevant in this respect that according to that Baraba Steppe paper, Andronovo and Iron Age mtDNA is not coming from Eastern Europe. For the most part, mtDNA is local, i.e. found in Bashkirs, Tatars and Volga-Uralic people, but the exotic haplotypes seem to have links to countries like Iran, Azebaidjan etc.
ReplyDeleteSome Baraba haplotypes are similar or close to the haplotypes of the following West Asian groups:
Andronovo Tartas:
T - Gilaki Iran
Iron Age Chicha:
H - Shungan Tadjikistan,
U1a - Azerbaidjan,
U4 - Shungan Tadjikistan
U5a - Hunza
T - Iranian Kurds, Shungan Tadjikistan, Ti Azerbaidjan
T1 - Bronze Age Kazakhstan kurgans, Kumandins, Mazandarians
J - Turkmen
H - Shungan Tadjikistan
U3 - Iranian Kurds, Gujarat
W - Mazandarians, Tadjikistan Ti
H6a1 - Hunza
It seems that only T2b, found in Baraba Chicha burials, is typically European and found in LBK and one Baraba Chicha U5a1 haplotype is found in Italy.
Maju: "This europid Z93* in the Altai"... "It is not "Europoid" in any way I can discern: just look at the haplotypes, for Chaos shake!"
ReplyDeleteWell as mentionnned, I was obviously referring to their morphology that the mainstream studies (references in the mainstream D. W. Anthony and J.P. Mallory's books as well) qualify as Europoid and more specifically as proto-europoid in east European studies.
The sudden appearance in south Siberia, around 3500 BCE, of Europoid population with early Yamnaya technology (objects, metallurgy), economy (pastoralism (also keep in mind the Mongolian cattle DNA being part "european")), culture (cultual object, kurgan) associated with west female lineages (with modern matches in Europe) and light pigmentation, really plead for a population movement from the west - and by west I mean the eastern part of early Yamnaya territory.
I can't just ignore the material archeological evidences.
"Isn't Central Asia "Western" (not necessarily meaning "European") since the very beginnings of the Upper Paleolithic? I don't need any particular explanation for Altai or other Central Asia aligning with Western genetics all the time before the Turkic migrations of the Iron Age: it's what I would expect considering its cultural links (Aurignacoid, Gravettian, Western Neolithic, etc.)"
As said, the female lineages happen to have European modern matches (as far as Iceland for the mtDNA H Tarim sample formerly mentionned), and some have very European-centered presence (for instance H5a (clearly European, not near eastern or Caucasian) in Kayzer et al 2009 and H11a (east European) found in a study about Udegeys - if I made no mistakes, besides haplogroups such as U5a1 (typically European), U4 or even some U1a associated with southern Russia and the "Maykop" region), etc...
No south Asian/south central Asian female lineages are found in the south siberian's most ancient aDNA.
The pre-neolithic pool of central Asian female haplogroups would have been quite similar to the recent European one and with no presence of south Asian lineages at all despite your surmised migration of R1a from there? Weird.
South Russia and the Altai are not close. If the Afanasevo people (end of neolithic) had early east Yamnaya axes and potteries besides Kurgans, they didn't come from west or south Asia.
"Also prolific farmers would be much more likely to cause a demic impact against the hunter-gatherer precursors than a bunch of Bronze Age raiders versus one of the greatest civilizations of that time."
I agree, but this doesn't seem backed at all by the south Siberian case. Clearly in this case it doesn't seem to have anything to do with such a process.
"the modal Z93* haplotype is quite divergent from the M417* ones"
I see no impossibilities here, though.
@ Kristiina about local mtDNA
Iron age is not really interesting to me because the mobility of Saka-like tribes allows for central Asian/south Asian haplotype appearance. I'm only interested by the oldest lineages.
It doesn't inform us much if many of these haplotypes you mentionned actaually have an ancient origin (or are derived) from ancient population movements from east Europe/Russia (As typically examplified by the bronze and iron age Kazakhstan kurgan's samples with T1 (Lalueza-fox et al 2006) - T1 is also present in north-eastern Europe IIRC BTW, or for instance with your U5a or H6a1 Hunza samples that have obviously an ancestor from somewhere else (IIRC H6 is present both in east Europe and central Asia). As for the presence among the modern Bashkir/volga-uralic haplotypes, I fail to see how it discards an ancient chalcolithic/bronze age origin or presence among antique eastern Kurgan populations).
"Europoid", whatever your usage, means European-like and in this case there's nothing European-like we can discern in Central Asian R1a.
Delete"The sudden appearance in south Siberia, around 3500 BCE, of Europoid population with early Yamnaya technology (objects, metallurgy), economy (pastoralism (also keep in mind the Mongolian cattle DNA being part "european")), culture (cultual object, kurgan) associated with west female lineages (with modern matches in Europe) and light pigmentation, really plead for a population movement from the west - and by west I mean the eastern part of early Yamnaya territory".
Unless we are missing something, right? Unless there is no "new population" and is just the same old herder-farmers adopting, after conquest a new identity. What do we know of the Neolithic of Central Asia, I mean: really, what do we know even of the Neolithic of the Samara bend! Who were the proto-Kurgan people?! We know almost nothing about the Neolithic of all the steppe, only since elites, gold and horses appear is when research resources become interested: gold calls gold, it seems.
"I can't just ignore the material archeological evidences".
Fair enough, I can't do it either. But DNA doesn't lie because nobody wrote the code: maybe we get confused interpreting the what, when and why but the DNA data is there and does not fit the steppe migration paleohistorical narrative.
It's also an interpretation problem for me, believe me, but that's what we have.
"... for instance H5a (clearly European, not near eastern or Caucasian"...
Whichever its exact origins, H5 seems deeply involved in Neolithic process in Central Europe, notably at the Tisza.
I really don't dare to judge Y-DNA evidence on mtDNA one. They may or not be related. It's perfectly possible that those mtDNA lineages (U5a, U4, etc.) precede the Y-DNA expansion and have Paleolithic origins in that area.
"I see no impossibilities here"...
I see what the Y-DNA data says. I would honestly prefer almost any other kind of data, because it'd help us to produce more easily a coherent narrative, but this is what the DNA says and only Nature wrote it.
We will have to solve the problem of putting the DNA facts together (along with other evidence) into a coherent narrative that should approach the truth better than anything else.
Maju: "Unless there is no "new population" and is just the same old herder-farmers adopting, after conquest a new identity. What do we know of the Neolithic of Central Asia, I mean: really, what do we know even of the Neolithic of the Samara bend"
Delete"Mediterranean"-like Farmers waiving and forgetting about their farming knowledge and way of life and transforming themselves into mesolithic europoid (general morphlogy, skulls) pastoralists and hunters, just as the ones found in Russia and Ukraine (_prior_ and after the emergence of the Samara and Khvalynsk cultures)? How is that credible?
" "Europoid", whatever your usage, means European-like and in this case there's nothing European-like we can discern in Central Asian R1a."
As of now, that's only an opinion. And we could say that there were a lot of europoid in the ancient population that was apparently likely a carrier of this haplotype.
I say that a population surmised to originate in Iran (mixing a bit in central Asia) that ends up with
1/ the same morphology mesolithic Russian/Ukrainian hunter-gatherers
2/ a sizeable north EUROPEAN autosomal component (found in several studies of the later years)
3/ typical (even specific) dental EUROPEAN characteristic (http://onlinelibrary.wiley.com/doi/10.1002/ajpa.21585/abstract). Precision: this double-rooted canine stuff is basically absent in the near east and middle east (it's present IIRC but very rare. Quite more than in the south Siberian region) and of course totally absent of south Asia IIRC.
This characteritic is found nowadays not only in south siberia but also in Mongolia, Tarim, ordos (Inner Mongolia, china), in all the region where the populations associated with these (sometimes partly admixed) proto-europoids were to be found, including in the Tarim Basin (with some other kind of caucasoid-like type during iron age IIRC, with the spreading of the Sakas (an even more admixed bunch apparently, not only with te east Asian type but also with a south and "west" Asian type))
4/ non negligible light pigmentation according to Bouakaze et al 2008 and Kayzer et al 2009 (in accordance with some pictures of modern individuals of these regions)
5/ to which we can add the presence of an "european" autosomal component in the cattle of this global region (we can assume it has something to do with the appearance of patsoralism in these region)
6/ typical shared technology between eastern Russia/Volga and south Siberia (specifically east Yamnaya-like)
7/ All this in a context where we can suspect a cultural and linguistic link between south-east Russia and south Siberia (tocharian) that happen to fit ith a mainstream theory (the Kurgan theory) explaining some common cultural and linguistic background for very different Eurasian populations.
... has actually NOTHING to do with Iran at all and unambiguously point towards the east of the Pontic steppes.
If your theory can't efficiently explain this, I dare say there's a hole in it.
"Fair enough, I can't do it either. But DNA doesn't lie because nobody wrote the code: maybe we get confused interpreting the what, when and why but the DNA data is there and does not fit the steppe migration paleohistorical narrative."
Indeed, DNA doesn't lie: Among west eurasian female lineages we have some european specific haplotypes (and more generally several modern matches to the ancient samples in europe, in diverse studies - including a Chinese one) and a sizeable north European component (also seen in different studies). You said it: DNA doesn't lie.
You don't actually _KNOW_ where is the origin of the Z93 branch with this study, we can only surmise. Is there an _impossibility_ in the fact that it would have originated roughly in the east of the Volga, which would fit nicely with the Kurgan theory that you happen to support so far?
"Whichever its exact origins, H5 seems deeply involved in Neolithic process in Central Europe, notably at the Tisza."
DeleteYou're talking of H5 wich has a wider spread. I was specifically talking of H5a which is European-only (like the H11a in the udegeys I mentionned, which is there among other west eurasian mtDNA hgs, here (behind a paywal) http://onlinelibrary.wiley.com/doi/10.1002/ajpa.21232/abstract). and was found in the south Siberian aDNA in Kayzer et al 2009.
I agree that U5a1 and U4 (and U2e) might have arrived there independantly earlier, even if U5a1 at the very least seem really originally European-specific - no certitude though, they could easily have been embedded in a chalcolithic movement too.
Re H5a: it can well repesent anything, even Paleolithic connections with Europe (for me mtDNA H is very old and certainly pre-Neolithic, so if U subclades could link Europe and Siberia in the Gravettian, so could H subclades as well).
DeleteRe. morphology: when you talk of European or Europoid, I assume that you are well aware that there are no obvious differences in those elements with West Asians: you can't decide with any certainty if a skull is European or West Asian: it'd be a coin toss. You can't discern the skull of Jamenei from the skull of Mdevev, for example, you can't discern the skull of Erdogan from that of Schröder, even Egyptians and Norwegians have nearly identical skull shape! There's nothing obvious segregating Europe from West Asia in phenotype.
Re. pigmentation: we know very little about pigmentation genetics yet but, in any case, the alleles usually associated with lighter skin are in essence a Neolithic import, so they should have arisen in West Asia, not Europe.
"If your theory can't efficiently explain this"
I do not have a "theory" (not yet): I just have data and conclusions forced by that data. Even if you'd be right in all your very oblique arguments, that wouldn't change a comma about how the R1a geo-phylogeny actually is. It would be contradictory evidence but wouldn't be able to dismantle the Y-DNA factoids.
What happens when you have contradictory evidence? Either you can prove some of it wrong in its own terms or you have to find a solution that conciliates that apparent contradiction.
Beating a dead horse is useless: it won't get up ever.
"You don't actually _KNOW_ where is the origin of the Z93 branch with this study"...
I'm pretty sure I do. As I said before: please check the damn haplotype structure tree: all main branches stem from the South or have the majority of their early branches in the south. That is totally inconsistent with your hypothesis of a Northern origin.
The authors also confirm that the Siberian branch is apparently derived and not ancestral:
... lower diversities occur in south Siberian paragroup R1a-Z93* (H¼0.921), in Jewish R1a-M582 (H¼0.844) and in Roma R1a-M780 (H¼0.759), consistent with founder effects that are evident in the network patterns for these populations (Supplementary Figure 2).
So Underhill et al. and I are in the same page in this aspect. Just that I write a blog and they write a scholarly paper, so we use different forms of expressing the same fact.
"Is there an _impossibility_ in the fact that it would have originated roughly in the east of the Volga (...)?"
Yes, it is a practical impossibility. Only terminal branches are found in Europe in fact (excepting the Roma).
I admit the matter at this point is blurry but I have to react to a few points:
Delete- You are the only person I know that would allege _mesolithic_ european hunter-gatherers were undistinguishable from south or west Asian populations. There WAS certainly a clear distinctive morphology between the south-east European neolithic farmers and the Ukrianian/Russian (and south Siberian) hunter-gatherers - then pastoralists.
The specialists are clear about that.
- You are the only person I know that would allege that the sizeable north European component in autosomal data in south Siberia is representative of a mostly south Asian/west Asian population.
- You are the only person I know that would allege that the _high percentage_ of light-pigmented eyes and hair among the individuals of ancient south Siberia (aDNA of Bouakaze et al 2008 and Kayzer et al 2009) fit the profile of population mainly derived from a south/west Asian population.
About pigmentation in ancient populations, we only have partial data so far. The full association of light pigmentation with west Asian neolithic newcomers is very flimsy, especially since the tested individuals in mesolithic Sweden are not representative of the current - particularly light-pigmented - population of this region: The Y-DNA hgs are completely different from nowadays even if they are derived from the same ancestor (hg I).
Y-DNA I1 is the haplogroup to which there is a particular correlation with light pigmentation in modern European regions, not I2, and this hg wasn't part of the tested mesolithic population - which implies population disparition/replacement.
And some light eyes were found among these European mesolithic samples and not in the neolithic farmers ones, so let's not affirm anything before every corner of pre-neolithic Europe is tested and every haplogroupfound.
I mean, if the south Siberian Z93* population is from an ancient mix of paleolithic Iranians and paleolithic European-like hunter-gatherers, where is the "European-like" Y-DNA hg (hg I?)? - the ancient Kurgan male lineages were fully R1a.
Why no south Asian mtDNA hgs at all. Why no south Asian autosomal component (or very few, I don't remember - in this case, it can be associated with Saka-like input, at this level)
"Yes, it is a practical impossibility. Only terminal branches are found in Europe in fact (excepting the Roma)."
DeleteThat doesn't say anything about the origin of M417, especially if we accept your favored molecular clock estimates - it "strongly suggests", if you will.
Particularly since we did actually find some ancestral R1a lineages in Europe, even if not numerous.
Ancient lineages do vanish (where are Otzi lineages in the region he was found? Where are the ancestral stages of the quite derived Japanese Y-DNA D2? They're gone. Impossible to know where this D2 appeared at this point) and recent unexpected aDNA results taught me to be cautious about haplogroup history and that the obvious scenario is not always the right one.
If you cling on the European R1b being present in paleolithic Europe with the major part of mtDNA H despite a lack of positive samples in paleo/mesolithic samples (except maybe some H but the signal is faint, especially far back in time), I think I can say that R1a-M417 in Europe is not to be completely discarded.
"The authors also confirm that the Siberian branch is apparently derived and not ancestral"
I never alleged the Siberian Z93 was the ancestral root. It's still not within Z2125 or M780 (notice how these larger haplotypes on the maps are souther than the derived - but still a Z93 not within Z95, If I'm not mistaken - south Siberian.
As for the Jews and the Roma they are "little" communities that are quite endogamous, it might have helped to preserve old lineages.
Besides this author also claimed in 2009 that India was the source of R1a because of its particular diversity there. Well, we know he was wrong, even by a mere look at the R1a tree, so... he's not infallible :)
Anyway, one sure thing, a south Siberian Z93 coming from south Asia would have to have arrived in south Siberia as ancient hunter-gatherers way before neolithic, otherwise it wouldn't add up.
"You are the only person I know that would allege that the sizeable north European component in autosomal data in south Siberia is representative of a mostly south Asian/west Asian population".
DeleteI don't believe I said that. I tend to consider autosomal data and haploid data separately: they tell us of different things. Autosomal data as analyzed with Admixture and similar tends to give shallow (recent or sometimes even just wrong) results unless you go quite deep in the K levels, close to optimal cross-validation ones, typically >15 in subcontinental samples, many more when global. Consciously or unconsciously it can produce wrong or highly misleading results.
Haploid DNA is more straightforward.
"You are the only person I know that would allege that the _high percentage_ of light-pigmented eyes and hair among the individuals of ancient south Siberia (aDNA of Bouakaze et al 2008 and Kayzer et al 2009) fit the profile of population mainly derived from a south/west Asian population".
The Lazaridis data is clear re. the (known) light skin color alleles being only present among EEF (early European farmers) and not WHG (Western Hunter Gatherers). The opposite is true about the eye color allele if I recall correctly. There's a lot of people who believe that those skin color alleles have anyhow been selected for only very recently (I have no clear stand on that).
"The full association of light pigmentation with west Asian neolithic newcomers is very flimsy"...
I am the first one to argue for that weakness of the genetic evidence, because a lot of the skin and hair pigmentation genetics are simply not known yet. BUT the allele distribution was clearly slanted in favor of the EEF group, regardless of their effect on actual skin color. They are genetic markers no matter their effect.
In the same line there was recently some noise about alleged "dark skinned Kurgans", although I admit I did not pay too much attention (sorry, no time). See:
→ http://dienekes.blogspot.com/2014/03/dark-pigmentation-of-eneolithic-and.html
"... especially since the tested individuals in mesolithic Sweden are not representative of the current - particularly light-pigmented - population of this region: The Y-DNA hgs are completely different from nowadays even if they are derived from the same ancestor (hg I)".
Precisely: are we talking of past populations or present day ones? You seem to be happily mixing both without making much sense.
"I mean, if the south Siberian Z93* population is from an ancient mix of paleolithic Iranians and paleolithic European-like hunter-gatherers, where is the "European-like" Y-DNA hg (hg I?)?"
I don't see any reason to expect I in Altai: Mal'ta 1 was R* and we know that proto-Amerindians who migrated Eastwards from there carried Q1. I2a seems some sort of European specific lineage. Altaians have sizable frequencies of Y-DNA Q1, as we should expect their Paleolithic heritage to be.
Paleolithic Europeans must have got different (albeit related) founder effects to those of Central Asia (Altai included): both founder populations migrated from the same origin (West-South Asia: somewhere in the Delhi-Thaskent area very possibly) but almost certainly separately and with only limited contact thereafter. They belong to the same West Eurasian macro-population but that's about all the relationship that Ma1 has with Europeans (excepted some minor flows/admixture concentrated in Eastern Europe and possibly brought to the West by Indoeuropeans, who did not bring much R1a over here anyhow).
BTW, if you want to ponder Central Asian and West Siberian autosomal DNA, you can't skip this:
Deletehttp://leherensuge.blogspot.com/2010/07/central-eurasian-genetic-specifity.html
The much ignored Hui Li paper found a Central Asian/West Siberian specific autosomal component which is half-way between West and East Eurasia by Fst distances. It is maybe a mixed component which includes almost surely Ancient Siberian (closer to the West) and some Siberian inputs.
In any case, once accounted for, the European influence in the area collapses to near zero, while the East Asian influence (Turkic) remains important.
This is not the Ma1 "ANE" component because it has very low influence in Europe but it is almost certainly related to it very closely (autosomal components are often fluctuating, as they are just measures of affinity, not absolute things).
"That doesn't say anything about the origin of M417, especially if we accept your favored molecular clock estimates"...
DeleteThe molecular clock estimates are irrelevant in the localization of the approximate origin. That depends only on the geographical structure of the phylogeny, particularly its most basal diversity.
"Ancient lineages do vanish (where are Otzi lineages in the region he was found? Where are the ancestral stages of the quite derived Japanese Y-DNA D2? They're gone. Impossible to know where this D2 appeared at this point)"...
Vanished lineages are pointless to this discussion, I believe: we're talking of existing lineages and their informative role on the origins and spread of the very real set (haplogroup) R1a.
As for D2, we can know that D originated near Burma or Yunnan (highest basal diversity is over there) and that D2 is an offshoot. D2 may well have coalesced already in Japan or not far away. It is true that we can't infer the exact route of the D2 founders but that is not that important, is it?
"If you cling on the European R1b being present in paleolithic Europe"...
I don't "cling" but I can't reject it either. Some data suggests so, other is less supportive.
"(except maybe some H but the signal is faint, especially far back in time)"
The signal is the signal. It does not depend on time but rather on resources spent in actually testing for it.
"I think I can say that R1a-M417 in Europe is not to be completely discarded".
I don't see any evidence of it. Certainly not with the data of this key study.
The M417 expansion appears to have been fast sending some "asterisk" elements to the North Sea in the process but otherwise nothing points to any European geography at the origin nor in the route: it seems clear that the center of the expansion was towards Iran or somewhere nearby. One of the two main subcenters was European: that of Z282, but that's about it.
Of course: if new data that contradicts this comes around, then I will (again) move my stand accordingly, always trying to follow what the data says.
...
...
Delete"but still a Z93 not within Z95, If I'm not mistaken - south Siberian. "
If you look at the haplotype structure (please do!), Z93* has the following structure (names are arbitrary):
→ A Caucasus (singleton)
→ B Middle East (singleton)
→ C:
→→ C1 South Asia
→→ C2 Altai
→ D:
→→ D1 South Europe
→→ D2 Middle East, South Europe, Altai (one sub-branch each)
→→ D3:
→→→ D3a Caucasus
→→→ D3b:
→→→→ D3b1: South Asia (singleton)
→→→→ D3b2: three branches with basal nodes in South Asia and derived groups in various places (incl. Altai but not only)
So it is South Asia → Altai and not the other way around. This migration may be a bit older than the rest of Z93 but it still stems from the South and has Altai in the end, even if frequency may mislead.
"As for the Jews and the Roma they are "little" communities that are quite endogamous, it might have helped to preserve old lineages".
What helps them is to have very specific founder effects, what reduces their diversity. Both communities have clearly expanded since their founding in Medieval and Early Modern times, so there is little pressure to reduce diversity: their reduced diversity is clearly caused by founder effects instead. The same happens with Altaians, which seem to have exactly five Z93 founder effects: very specific founder lineages derived from the South (one of them is ambiguous but all the rest are not).
"Besides this author also claimed in 2009 that India was the source of R1a because of its particular diversity there".
Was it Underhill? I though it was an Indian researcher.
"... he's not infallible"...
Nobody is. You neither.
"... a south Siberian Z93 coming from south Asia would have to have arrived in south Siberia as ancient hunter-gatherers way before neolithic, otherwise it wouldn't add up".
Why not? What do you know of the Early Neolithic of Altai. I don't know anything but I do know that further South in Central Asia, the Neolithic arrived from West Asia, as everywhere else in the wider macro-region (i.e. excepted East Asia and America, where it evolved autonomously).
I must say, BRAVO for this piece Maju. We are plagued by internet pseudo-analysts and their ethnocentrism (Steppe theory). This piece regarding R1a origins makes it the truth glaring - R1a and specifically Z93 came from West Asia, and likely Iran. Steppe theorists have been indulging in their delusion for too long.
DeleteUpdated with my molecular clock estimates for all the fig. 5. All based on age(CF)=100 Ka ago, as archaeological evidence strongly implies. IMO R1a-M417 expansion in Europe as in Asia seems Epipaleolithic in essence (a quite fast one in any case), R1b instead looks older and in Europe likely "Magdalenian" (whatever reshufflings happened later on).
ReplyDeleteThere are North American specific R1 lineages too aren't there? How would they fit into the phylogeny shown in your post?
ReplyDeleteI don't think there is any Native American R1 that cannot be attributed to colonial admixture (please tell me if I happen to be wrong). However long ago (2006?) I read something (older) about some R* in NW Native Americans and also separately about possibly (but unclearly) related Mongol R* too. The studies must be very old, 2004 or something like that, and in a recent search I could not find them online. It'd be interesting to "rediscover" them on light of the recent Mal'ta "missing link" data.
DeleteOn second thought and after some new search, it seems that I was ill-recalling the matter and what the old studies actually found was P(xQ) (→ http://www.ucl.ac.uk/tcga/tcgapdf/Bortolini-AJHG-03-YAmer.pdf), whose high frequencies among some Native Americans, notably the Chippewayan (63%) are odd to say the least. Other populations include 0-21% of P(xQ).
DeleteThe most common and quite natural attitude on that data is to discard those percentages as belonging to colonial admixture of European origin (i.e. R1b/R1a). However I'm pretty sure that other studies (which one?) suggested it was partly P(xQ,R1). In that time P* and maybe even R* was not yet reported so we speculated with R2, I recall, but that also made no sense in that geography.
Recapitulating now it's probably P* or R* (Mal'ta like?) but I can't confirm anything at this point. The issue has not shown up in more recent studies to my knowledge, so either there is a tendency to discard all P(xQ) as being "recent European" (very possible) or the sequencing methods of that time could not properly identify the lineages (less likely IMO).
I found something else:
Delete→ http://mbe.oxfordjournals.org/content/23/11/2161.long
→ http://mbe.oxfordjournals.org/content/21/1/164.full
All R found among Native Americans seems to be of recent European origin. However there is still some P(xQ1,R) which may well have older roots (no obvious European association found). Notice that this P* can also be Q* but not R in any case.
Here's one source http://www.sjdimond.us/M3%20ancient%20links.pdf - it has R1b1b1-M73 prescent in Sibera back 18.2±10.5 Ka but states that further SNPs need to be identified to infer more about the migration history. I'd swear I've seen other papers on this two but I can't seem to find them now for the life of me.
DeleteI should have been clearer that I meant the R haplogroups found typically in speakers of Algic (or more specifically Algonquian) languages - people like the Chippewa/Ojibwe, the Cree, and so on.
Unhelpfully, these are also the peoples with some of the longest and most extensive contact with Europeans, first with Basque fishers, then with French and later Scottish and English fur traders. So I'm sure there's admixture too. Unhelpfully, the least admixed populations are also the ones least likely to get sampled. Conditions on the more isolated First Nations reserves in Canada are absolutely deplorable.
My issue with these older studies is that because they predate the work on Mal'ta, they tend to assume that anything with European affinities in indigenous people in the Americas must be colonial in origin. We now know that Ancient North Eurasians were a thing, and that Europeans and indigenous people of the Americas have the closest affinity to them today. European-Amerindian affinity does not necessarily mean admixture in the last 500 years. Interestingly, that Bortoloni papers shows Algonquin peoples (Chippewayan and Cheyenne) as having the most affinity to Siberia, as do the Yanomami in South America. Alongquins and Yanomami are both conspicuous in having high levels of mDNA haplogroup X2 as well, which I don't think can plausibly be attributed to admixture. In Eurasia, haplogroup X2's frequency peaks right where you place R1a's origin.
Just a general comment here too by the way, but it seems to me that far too much emphasis is place geography and far too little on ecology when it comes to archaoegenetics. I'd note that if you look on a map of vegetation from the last glacial maximum, the area where R1a was savanna. The next closest area with that type of vegetation was in NW Africa - the likely origin of R1b's expansion in Europe and Africa.
Have you read the links I searched for? All sampled Native American R1 haplotypes are derived from European ones. If you think otherwise: get the haplotypes and compare them with Siberian ones. Then send me the results by email or publish your own study or blog entry.
DeleteMal'ta 1 was R* but he was already a leftover of the proto-Amerindian migration, which was by then in the Russian Far East (or somewhere nearby, clearly East of Lake Baikal), maybe even already in Beringia.
The timeline of the UP expansion to NE Asia (Mongolia, North China) begins c. 30 Ka ago and that is almost certainly the time of the proto-Amerindian migration to the Far East: some 6000 years before Ma1.
It's only logical that each of the populations fixated their own P-derived lineages, most commonly Q variants, but that for some reason Ma1 still retained some P-other, specifically R* (which was not yet R1, nor R1a, nor R1b nor R2 but just a distant "uncle" or "cousin" of these).
Autosomal DNA still allows us to identify Ma1 as closest to modern Amerindians but his uniparental lineages were not particularly close to them anymore (founder effects, fixation... the inevitable effects of separation in time and space). R* specifically seems close to nobody in particular: it's just another P-derived Western lineage like R1 or Q.
I thought I'd clicked on both but apparently only the one. Sorry.
DeleteThis one (http://mbe.oxfordjournals.org/content/21/1/164.full) doesn't really sample the Algonquian peoples I'm talking about, with the exception of the Cheyenne. I don't disagree with the conclusions - I'm sure the vast majority of R1 in North America is from admixture - it's just the areas with crazy high numbers that I'm referring to. This paper also uses some REALLY out of date classifications for language families in North America. Algonquian and Quechua have about as much of a discernible relationship as Basque and Iranian or wherever else. The relationship may be there if you go deep enough, but assuming one is only assumption
The other is a lot better, and it looks like you may be right that I am thinking of P-M45* and not R1 (or people were reporting P(xQ) as R). I was digging through the Bortolini paper above just now looking at this and noticed the people in the town they got the largest sample for the people they are calling Chipewayan are actually Dene and Metis. Really Chipewayan people are Algonquian speakers, not Dene speakers. Metis is the Canadian term for descendants of European fur traders with Native American (well, Canadian First Nations is the proper term) wives. I don't know if they were able to separate Metis people out or not, or if there's even any real structure between the self-identified Metis and Dene people there. :/
I'll do more reading when I can. I'd swear I'd read solid sources on this. Or plausible at least. You would think people who are hunting mammoths and reindeer would be extremely mobile, so I don't think some sort of later admixture into Berigina after the initial settling is completely out of the realm of possibility. Hrm.
Please note that the Chipewyan (Denesuline) people are a Dené- (Athabaskan-)speaking people of northern Canada. The Chippewa (Ojibwe, Ojibwa, Ojibway, Anishinaabe) people are an Algonquian-speaking people of southern Canada and the northern United States. The Chipewyan and Chippewa ethnonyms may appear a bit similar to each other, but they refer to two different ethnolinguistic groups.
DeleteMuch needed clarification. Thanks, Ren Doe.
DeleteIt is the NE Dené Chipewyan people who showed the 25% P* in the Bertollini study. They are the only Na-Dené population in that sample, so they stand out a bit for both reasons.
Ah, yes I was getting confused there. Thanks. Though based on this http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3761611/#!po=32.1429 the Chipewyan apparently cluster with Algonquians genetically (and historically were allied with Algonquian groups against other Dene). So maybe the dinstinction isn't as important as I thought (and Bortolini's Dene sample was from a population not typical of other Dene). It would be nice if the Cheyenne sample had been separated out as well rather than just lumped with other "Amerind" groups.Europe
DeleteI'd suggest taking a look at that paper above. It's really quite interesting. They argue for at least two post-Clovis migrations. The first is dated to~8-10 Kya and associated with mDNA groups X2a and C4c. It peaks in Algonquian speakers but is present throughout Northern North America.
The second is dated to 4 Ka and associated with mDNA haplogroup A2a. It probably originated in SW Alaska and spread to Asia, Northern Canada and Greenland, and they attribute it to the Arctic Small Tool Tradition. It later (around 1000 AD) expanded to the SW United States with the Dene speaking Navajo and Apache. It is not found at high levels in groups like the Algonquian speakers.
The X2a / C4c are the ones I find particularly interesting, given C4a and C4a'b'c's presence in kurgans as early as ~6ka. The C4a'b'c breakup is still pretty old (http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3006427/figure/pone-0015214-g001/ is saying ~20ka) with X2 splitting up at a similar date (http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1180497/#!po=25.0000) but they are still very interesting haplogroups (particularly with X's roots in the Near East).
I don't believe a Y chromosome group has been identified that corresponds to the migration yet. This P* does seem like a likely candidate though. It would be really interesting to see how it fits into the overal P/Q/R phylogenic tree, and where related P* groups could be found.
It would be interesting to see a close look on the European-derived Y lineages too really. I would expect there would be quite a lot of variation based on the date of contact and the location. For the Mik'mac you'd expect a lot of Basque and French. With the Assiniboine you'd potentially see more of a Scottish/French mix.
By the way Maju, you may be interested in knowing that the Algonquian-speaking Iroquois' ethnonym has a Basque etymology proposed for it, via Basque-Algonquian pidgin. Supposedly comes from ilo kuo? "Killer People?" They were the enemies of the Mik'mac on the coast apparently. http://www.ehu.es/ojs/index.php/ASJU/article/viewFile/9275/8503
Apparently that error I got when I tried to reply earlier meant it didn't go through lol.
DeleteThanks Ren, that does help.
I did a bit more reading, and it seems that Chipewyans cluster more closely with Algonquian speakers anyways, and were traditionally allied to Algonquians against fellow Dené, so the distinction might not be as important as I thought.
Based on mDNA (see http://www.pnas.org/content/110/35/14308.full.pdf for example) people have proposed two major post-Clovis: one at around 10kya associated with C4c and X2a, and another from Alaska or the Chukchi Peninsula associated with A2a and the Arctic Small Tools tradition dating from around 4.5kya.
C4c and X2a are mostly specific to the Northern half or two thirds of North America, and are most prevalent in both Dené and Algonquian speakers, and virtually absent everywhere else (with a couple of exceptions).
A2a is specific to the Dené, the Inuit and a few associated groups in NE Asia.
It's the C4c/X2a group that I find interesting in this context. What Y DNA groups were associated with this? This P* sounds like a plausible candidate. Can relatives of P* be found Asia? How does this P* branch fit in with Q and R?
X2 seems to have originated in the Near East around 20,000kya (http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1180497/#!po=15.0000), which is pretty interesting in and of itself. C4a'b'c seems to have split up around the same time. What is most interesting to me though is that C4a'b'c and C4a were found at very high frequencies in the early Kurgan remains.
What's the Y-DNA side to this story? How does it tie in with the broader history of northern Eurasia?
Sometimes Blogger fails to send me an email of comments waiting for approval and when I get to check there is one or several awaiting. This time I found a bunch of them. My apologies about that.
DeleteNo worries lol. It did actually give an error on my end too at the time. I'd love to hear your thoughts whenever you get the chance. Cheers.
DeleteRe. mtDNA molecular clock estimates: they can well be very wrong (I often almost double them to make any sense, but it varies). I would not elaborate too much based only on such feeble estimates, unless there is other evidence. It's possible that the 8-10 Ka estimated clades are Clovis for example.
Delete"Can relatives of P* be found Asia? How does this P* branch fit in with Q and R?"
P* just means P-other, normally P(xQ,R). It's most common around Bengal but the lineages found over there could well be as different from those found in North Asia or America as Q is from R. Only a detailed haplotype study could shed light on this issue and AFAIK there is none.
"What's the Y-DNA side to this story?"
Not sure how to interpret this question. X2 seems among Siberian and Amerindians the female counterpart of yDNA Q1 (which also originated in West or South Asia). However while Q1 remained dominant, thanks to patrilocality, X2 and whichever other Western lineages originally associated with Q1 shrank to almost nothingness, being replaced by East Asian lineages (A, B, C and D). A similar but inverse pattern we see with N1 peoples of mostly Uralic ethnicity, where the Eastern Asian mtDNA lineages have almost (but not totally) vanished in spite of yDNA N1 being still very dominant. In general lines, at least in these two cases, autosomal genetics follows the pattern of mtDNA and not or very weakly that of Y-DNA.
Re. the "Iroquois" as Basque-pidgin name, I find the paper convincing in the part of the common ethnic suffix -quois being probably Basque -koa originally (i.e. "that from..." whatever precedes it. "Bilbokoa" form example means "the one from Bilbao", "Frantziakoa": "the one from France", etc.) This is not really genuine French, although a similar suffix -ois does exist in this language with similar meaning AFAIK, so Québécois can be interpreted to be genuinely French (Québéc+ois), much as François is as well (France+ois). Probably even the evolution -koa → quois reflects this colonial mixed evolution, somehow retaining the Basque -ko- but adding to it (rather redundantly) the French -ois.
DeleteSo far so good. But I can't really agree with the prefix Iro- meaning "hil(-du)" (to die or to kill, notice by the way the similitude between the Basque and Germanic words, which I consider cognates, probably Vasconic substrate influence into Germanic). Basque at the very least does not confuse /r/ with /l/ ever, nor does French (even if their "r" sounds like gh). Instead I would search for a more likely equivalent to iro- or hiro-
First I though of "hiri", which means city or town, but I don't think that Iroquois settlements were so populated nor that the i→o vowel shift seems likely either (they are opposite in the vocalic triangle).
Instead what about "hiru", which means three? Three were the matrilineal clans of the Mohawk, and AFAIK of all the Iroqois tribes: bear, turtle and wolf. Maybe that's what Iroquois means: hirukoak: the ones from the three (clans).
PS- A possible reason to the retaining of this pidgin Basque form could be that the Iroquois originally lived in Eastern Canada (Gaspé peninsula and New Brunswick) but were expelled by the Micmac in the 16th century. In fact the name Gaspé comes from a Micmac exonym for the Iroquois or Mohawk: Kwedech. By that time Basque fishermen were already in the area (since at least 1512). It seems plausible that French fur traders first (since c. 1580) and settlers later (since c. 1603) retained Basque ethnonymy for many of the nations of the region.
DeleteTown-people could be plausible. The Micmac were more nomadic than most Iroquois weren't they? The Micmac relied more on hunting and fishing while the Iroquois mostly farmed corn, beans and squash.
DeleteThere were a lot of population movements due to the arrival of Europeans on the Atlantic Coast. I think that's part of what makes it so interesting. The development and spread of new technologies in Eurasia caused huge population movements too, but it's rare that we have such good historical records of it happening as we do with firearms in North America.
These migrations are how the land my father's family settled on become vacant. As the Iroquois moved West, they came into conflict with the Huron, with the Huron not faring very well in the exchange. It is in these depopulated Huron lands that my Scottish ancestors originally settled. Worked out well for me I guess. Not so good for the Huron unfortunately.
Re: P* - yah, I realize that just means P(xQR). I think such a detailed study would be very very interesting. Some P* groups would presumably be related to the Chipweyan P*. Some wouldn't.
Yah, X2a and C4c could well have arrived at the same time as B2 and others. The structure is quite striking though. I think at least an arrival by a different route is the most likely scenario, even if the two migration routes happened simultaneously. If there were two different groups migrating with two different mDNA profiles, but with the same Y-DNA, why and where did this structure form?
Or, if there really was only one migration route why did X2b and C4c dwindle to almost nothingness in South and Central America but not in North America?
Q1 is almost 100% in South America and X2 is virtually 0 there. In fact, Q1 has its lowest prevalence in North America precisely where X2 peaks. They're anti-correlated.
Anzick-1 has a much closer affinity to South American populations than to Algonquian and Dene groups, even those that live directly adjacent to where he was found. From the abstract of the original Anzick paper (which I haven't found in free form yet): "Finally, we find evidence of a deep divergence in Native American populations that predates the Anzick-1 individual." When did this divergence occur? How? Why? Presumably when you migrate into terra nullis, that has to involve both men and women migrating together.
In terms of the timelines - the timelines for the migrations seem sensible, don't they? I think the expansion of the Arctic Small Tool Tradition is fairly well dated, and it corresponds well with both the date and the distribution of A2a.
For dating C4a'b'c and X2... yah, granted. Here's the paper where I'm getting the C4a'b'c breakup from (http://www.plosone.org/article/fetchObject.action?uri=info:doi/10.1371/journal.pone.0015214&representation=PDF). The dates they quote seem to more or less agree with yours when it comes to Siberia and Beringia. C4a'b'c is really the most interesting one to look at IMHO given its presence in northern North America and in early kurgans. It seems to suggest that the very first Indo-Europeans may have had some fairly deep Siberian affinities. Here's the molecular clock they were using:
Values
of mutation rates based on mtDNA complete genome variability data
(one mutation every 3624 years [19]) and synonymous substitutions
(one mutation every 7884 years [19]) were used
Notice please that the P* among Amerindians can still be Q(xQ1) and not P(xQ,R). As far as I could find, it is P(xQ1,R), so it can perfectly be Q* (and I will suspect so until proven wrong).
DeleteAs for "hiri", notice that for Basques a town would not be the same as a village ("herri") but a large settlement, possibly walled in those times. For example here in Biscay there were two types of territorial organization: the towns (which had no vote in the regional parliament and applied royal law) and the so called "plain land", which was divided in republics (aka churchyards, because eventually the community meetings took place before churches). All towns were your usual medieval walled and compact settlements. These republics often had one or more villages as well as many scattered farmhouses. They were familiar with European Medieval urbanism, so my question is would Basques or any other European call "town" to a settlement made of wood and hay? I don't know but sounds a bit far-fetched. Instead the concept of hirukoak sounds perfectly good to me, and the phonetic change is very much minor to present day Iroquois (once we accept the French -ois addition).
Of course there could be an Iro or Hiro place from where they took the name as well, which would be Native, not Basque. That we may never know.
"Q1 is almost 100% in South America and X2 is virtually 0 there."
What corresponds perfectly with a founder effect, something we should all expect: as the population expanded through a narrow corridor (Central America) the diversity decreased. The most common lineages had nearly all the chances to make it, instead the rare ones had very small chances. Y-DNA Q1 managed to remain dominant because of patrilocality but for that same reason mtDNA X2 got diluted to near nothingness only surviving in some areas because of great luck.
"Anzick-1 has a much closer affinity to South American populations than to Algonquian and Dene groups"...
Actually the closest living population to Anzick-1 are the Maya, a North American population. South America begins only around Panama.
If you need a copy of the Anzick paper, I can send you one. I will need an email though.
"When did this divergence occur?"
Either in Beringia or at least North of Montana. It's possible that the Dené represent a secondary Siberian (not Beringian) migration anyhow.
"Presumably when you migrate into terra nullis, that has to involve both men and women migrating together".
Certainly. But the proto-Amerindians have a long paleohistory before reaching Beringia: they migrated first through Siberia and quite probably even Mongolia and North China (Q has been found over there with Neolithic ages) between c. 30-20,000 Ka ago, spreading with them the "mode 4" or "Upper Paleolithic" tech in the East. In that period they experienced extremely strong East Asian admixture from the female side becoming the Native Americans we know (or at the very least their seed population). Only then they migrated to genuine "Terra Nullis" (Beringia and beyond).
" It's possible that the Dené represent a secondary Siberian (not Beringian) migration anyhow. "
DeleteThe C-M217 present in Dene groups almost certainly had to have come from Siberia. The A2a present Evenks, Koryak, Chukchi, Inuit and Dene people are very likely the result of a back-migration from coastal Alaska though. Perhaps an expansion of a mixed Siberian-Beringian population on the Alaska coast. There is structure beyond just this though. Also just to be clear, there are non-Dene speaking groups that show this affinity too (that occupy a similar geographic area as the Dene). Maybe para-Dene is a better term? I don't know.
"Actually the closest living population to Anzick-1 are the Maya, a North American population. South America begins only around Panama. "
I should have been clearler - I'm grouping Mesoamerica with South America. From the figures in that Anzick paper (which at least are publicly available) that seems like a valid distinction to be making. If this structure was purely due to a founder effect, we wouldn't see this contrast in the autosomal DNA would we? And you would expect that the bottleneck would occur at or close to the physical bottleneck - perhaps somewhere near the the Darien Gap in Panama. Instead, there seems to be a cline that starts in the SW of the US or NW of Mexico that runs to the North East.
Would it make sense to suppose that within Beringia whatever orientalizing process that was going on could have been incomplete? One group departs bearing mostly East Asian mDNA, and another departs at roughly the same time along a different route with still mostly Siberian mDNA?
I'll send you my email. I'd love to read it.
Re: Towns and the Iroquois - Hochelaga was a walled village with 3,000 people living in it when Cartier visited it in 1535. I'm not sure how that would have compared to Europe at the time. Re: the phonetic change, yah, that does make your scenario sound more plausible.
I wouldn't be surprised if some of the evidence that was used to support the Solutrean hypothesis was actually just from early Basque-Mik'mac contact. One paper I saw was comparing Mik'mac and Basque folklore, with some story about an owl that asks you questions and kills you if you answer them wrong being common to both. The paper claimed this was evidence of a 20,000 year old connection. It seems more plausible that this story was relayed from Basque fishers and traders to Mik'mac fishers and traders, and then through the Mik'mac back to the French.
"I'm grouping Mesoamerica with South America. From the figures in that Anzick paper (which at least are publicly available) that seems like a valid distinction to be making".
DeleteSeriously again: provide an email address and I'll send you a copy. I say because you obviously have some knowledge blanks and you also seem very interested, Ryan.
Let's be clear: NA means Canada and Alaska, SA means all the rest of America excepted the continental USA which is not considered in that study (and is probably intermediate at least to some degree). So NA only means, in principle, arctic and subarctic populations, many of which may have been in Beringia or nearby areas until recently.
"And you would expect that the bottleneck would occur at or close to the physical bottleneck".
There was a physical bottleneck between Beringia and the rest of North America until the end of the Ice Age. In those times the only migration routes were the coastal one and, intermittently, an ice free corridor in Alberta. The vast majority of Canada and large parts of the continental USA were frozen and effectively impassable.
Therefore the Anzick-like expansion (Amerindians or most of them) happened c. 17 Ka ago when those peoples reached Oregon or somewhere over there, which offered to them for the first time in millennia a "Terra Nullis" available for massive expansion.
There was probably another less marked bottleneck at the arrival to South America but that one was secondary. Possibly the remnant arctic populations later expanded somewhat but almost certainly not to the point to takeover of all North America (even if we exclude Mexico).
"Towns and the Iroquois - Hochelaga was a walled village with 3,000 people living in it when Cartier visited it in 1535. I'm not sure how that would have compared to Europe at the time".
Following N.J.G. Pounds, it'd be a middle sized town (2000-10,000 people). There were lots of them in Europe in the Late Middle Ages; he counts ~20 only in Northern France and the Low Countries. Most county level towns were that size in England.
So I guess you can consider that etymology too, although I'm puzzled by the undocumented shift hiri → hiro, which is rather unnatural.
"... some story about an owl that asks you questions and kills you if you answer them wrong being common to both".
Never heard of it. It actually reminds me of the Greek legend of the Sphynx instead.
Sorry for the slow reply. Just got to read the paper now.
DeleteRe: the owl/sphynx story - it's an owl in Algonquin legends and the sphynx in Gascon and Greek legends apparently. They argue it's a ~20,000 year old story with origins in the Upper Paleolithic. I'm skeptical. Found the paper - http://halshs.archives-ouvertes.fr/view_by_stamp.php?&halsid=55j2o0i5mmqf56jqo4n3347g85&label=EHESS&langue=en&action_todo=view&id=halshs-00734560&version=1&view=extended_view
"So NA only means, in principle, arctic and subarctic populations, many of which may have been in Beringia or nearby areas until recently."
In terms of the samples, more or less yah. I went back into the Reich et al paper to see where exactly the samples were from, and it's right at the subarctic to temperate woodland transition. The climate is unusually cold for the latitude in eastern Canada, but we're still talking about as far north as Kiev and Paris. Not exactly a frozen wasteland.
The Algonquian and Dene groups extent much further south than that. The Apache and Navajo are Dene and live in Arizona, so we're talking the same latitude as Cairo here. They show a close genetic affinity to the Dene groups in the Arctic too so I think it's reasonable to extent the conclusions to all or most Dene groups. There were also both Dene and Algic (the parent group of Algonquian) groups in California in historic times. The Cheyenne (which Reich et al identifies as "Inuit admixed" at a time depth of ~180 generations) are farther south than Gibraltar.
"Possibly the remnant arctic populations later expanded somewhat but almost certainly not to the point to takeover of all North America (even if we exclude Mexico). "
It's a pretty big chunk of Canada+USA we're talking about really. Here's a map of the language families of North America that's decent if you're interested. The languages marked Iroquois, Algonquian and Athabaskan are the relevant ones. Interestingly, the Pima and Yaqui (both Uto-Aztec speaking groups on the Mexican/US border) place somewhat between Algonquians and Anzick, perhaps reflecting some level of admixture and lending weight to the theory of their origins as mesolithic nomads in Aridoamerica rather than immigrant farmers originating further to the south in Mesoamerica http://theabysmal.files.wordpress.com/2012/06/255411_310921055668150_1837369544_n.jpg
Either way, it's a pretty big area we're talking about. Substantially larger than Europe as a whole. I'm not suggesting outright "takeover" or replacement of course. I'm suggesting admixture. The expansion and admixture didn't have to happen all at once either. The Algonquians were pretty involved in Eastern Agricultural Complex centre of domestication for one. There does seem to be a very noticeable genetic cline similar to the EEF/WHG/ANE clines in Europe, with the more northern groups showing more affinity to Siberia.
The reason I think this cline is so interesting is because whatever group contributed the "East Siberian" component to Northern North Americans (the term used in Reich et al), The reason I think that I find this so interesting is that it seems that whatever group that contributed ANE affinities to Europe had some "Eastern Siberian" affinities, as shown in the K9 and above in the Anzick paper. Perhaps a group somewhere between the Khanty and Yukaghir samples. Learning about Native North Americans might give insight into Eurasia too.
Link to Reich if you are interested and don't want to bother digging it up:
http://hal.archives-ouvertes.fr/docs/00/72/69/62/PDF/Reich_et_al_FullSubmission_2011_08_10709A.pdf
Cheers
"The Algonquian and Dene groups extent much further south than that".
DeleteBut they are surely not good representatives of the overall Native American peoples of the mainland USA.
Also the NA component decreases towards the East in Canada, so, judging only on that, the Cherokee could well be like the Maya in regards to Anzick affinity.
... "whatever group that contributed ANE affinities to Europe had some "Eastern Siberian" affinities"...
But that Eastern Siberian affinity is typical of all the Native Americans, right? Just that the post-Anzick groups of Canada show some extra Beringian/Siberian inputs, while the rest of Native Americans relate to them only at the same "first wave" node that Anzick represents.
Mal'ta relates with various other populations as follows (see first graph in this entry):
1. Native Americans.
2. Siberians which form a distinct line of admixture ANE-East Asia (but are not close to Native Americans).
3. Europeans and West Asians, with the geographic gradient we all know more or less.
So that they have some affinity to these or those Siberians does not automatically imply anything about their relation to other populations.
Also Na-Dene are in that graph slightly removed from mainline NAs towards precisely those Siberians who tend towards Native Americans and not towards those Siberians who tend towards Mal'ta nor mainline East Asia. Their "Siberian" component has nothing extra to do with Ma1 but with East Asia as such.
The Eskimo-Aleut on the other hand are like a varied mix of those Siberians who are most close to both Ma1 and other Native Americans, closing the "Siberian V" and making it a triangle with their diversity. So most likely only in the Eskimo-Aleut case there is some extra "ANE" introduced in Native America.
PS- Thanks for the Reich link. Does it relate to this discussion in any specific way?
DeleteReich tries to address population structure in Native Americans in a pretty thorough way, and also has a lot of information on the samples shared with both the Anzick-1 and Ma-1 papers. Unfortunately it's a bit out of date and doesn't include any ancient DNA. It looks like the final article was updated to include Saqqaq's DNA. The supplementary materials are available here: http://www.nature.com/nature/journal/v488/n7411/extref/nature11258-s1.pdf This figure is particularly interesting: http://www.nature.com/nature/journal/v488/n7411/fig_tab/nature11258_F2.html They weren't able to exclude more complex models though as their data was not of sufficiently high quality. Similarly, the precise relationship between Saqqaq, Dene and Inuit groups wasn't entirely distinguishable, nor were they able to say much about links between Eurasia and these more Siberian-aligned Native American groups. Reich specifically mentions Na-Dene groups as something to follow up on. Pages 22-30 were the most relevant I think.
Delete"Also Na-Dene are in that graph slightly removed from mainline NAs towards precisely those Siberians who tend towards Native Americans and not towards those Siberians who tend towards Mal'ta nor mainline East Asia. Their "Siberian" component has nothing extra to do with Ma1 but with East Asia as such"
This Na-Dene group seems to be visible in the Anzick study. I'm referring to the light pink group on page 8 of the Anzick paper. This Dene-like component peaks on either side of the Bering Strait as well as in Greenland Inuit but is all found all the way from Algonquin groups to Russians and Mordovians. Perhaps some very sort of mixing between proto-Uralics and proto-Dene in and around the Urals? Or am I reading too much into this? The admixture runs for Anzick don't include any African samples and include a lot more Native American groups than Davidski does (for obvious reasons - the Anzick paper is targeting a very different set of populations).
It's too bad that neither the Anzick paper nor the Reich paper include data for Ma-1. Maybe there's an update out there that does. Still digesting the Reich paper for now.
You may be interested in this figure that Davidski shared a while back (and who's source I'm unsure of). It shows the routes of dispersal of microblade technology from Lake Baikal. It would seem like a decent map of some Y-DNA R and perhaps Q groups. http://img18.imageshack.us/img18/5406/qcu6.png More related to the original R1a subject of this post - the Tepe Guran site is in the Zagros mountains. I think this is a decent explanation for how R or R1 made its way into the Zagros.
Also, I hope it's not confusing or anything that I prefer to use Inuit over Eskimo. Eskimo is a bit of a pejorative in Canada, but in the US my understanding it does not have this negative connotation. Names for groupings of aboriginal peoples in Canada are quite a bit different than in the US for a variety of historical reasons, not all of them very good.
You may be right about the Cherokee btw. The most eastern sample is from eastern Quebec (around Saguenay) but that doesn't necessarily tell us much about the genetic makeup of the southeastern US.
Dear Maju,
ReplyDeleteThank you for discussing this paper. Unfortunately I do not have access to this paper. Thankfully, I realised after coming to your blog that the Suppl. Material was free.
After going through it, I wish to put forward some issues. I hope you can shed some light on it.
1. I feel that there is a sampling bias in this study against South Asia which may have prejudiced the results towards the Middle East.
For example, 1765 samples were taken across Iran which has a population of 76 million. In contrast, only 176 samples were taken from Pakistan, a country with a population of 180 million. If we go by population to sample ratio, to get a fair estimate from Pakistan vis-a-vis Iran, a sample size of about 4000 should have been aimed at. So the no. of 176 samples is more than 20 times less than that. We ought to bear in end that in previous studies, Pakistan has been a candidate of high diversity for R1a1.
For the rest of South Asia i.e. India & Nepal, a total of 863 samples were taken i.e. not even half of the no. of samples from Iran. The combined population of India & Nepal is 1.25 billion or more than 16 times that of Iran and yet the combined sample from this region is not even half of that from Iran.
Out of the 863 samples, 387 were taken from Nepal (195) and the rest from Eastern India & 126 from Peninsular India - regions not known for great diversity of R1a1.
This leaves only 350 samples out of which 40 have been designated as Mixed, while 36 samples were taken from Central India. Only 274 samples were taken from North & North West India out of which only 127 samples were taken from NW India - another region of great R1a1 diversity ( as per earlier studies).
In contrast, more than 6,600 samples were taken from Europe, a region with a population of 740 million & approx. 650 samples were taken from Turkey which has a population of 74 million.
In this scenario, how do we expect a fair assessment of R1a1 diversity within South Asia ? I would like to read what you have to say on this.
2. For a large no. of samples from South Asia and Central Asia, table S3 shows haplogroup M576. Yet this haplogroup/haplotype is not mentioned anywhere else. Can you throw some light on this ?
3. Finally a minor point I think which is nevertheless important. Afghanistan has been considered a part of Central Asia which in my opinion is not correct. Afghanistan has been politically since the very earliest times been part of South Asian empires, especially the region South of the Hindu Kush. Even during the Harappan phase, it was connected with the South Asian cultural sphere. Genetically too, the y-dna of Pashtuns (the largest ethnic group of Afghanistan) is closest to Indians and Pakistanis. Pashtuns also have the most homogenous Y-dna profile among Afghan ethnic groups which suggests very little y-dna introgression in comparatively recent times, including from India/Pakistan. This indicates that atleast the y-dna heritage of Pashtuns which is shared with the South Asians, goes back many millenia. It possibly goes back to pre-Harappan times and reflects a common origin of these people.
"Unfortunately I do not have access to this paper".
DeleteI can send you a copy if you provide an email.
"I feel that there is a sampling bias in this study against South Asia which may have prejudiced the results towards the Middle East".
I can't deny that you seem to have a point. I guess that Indian legal restrictions to DNA research may have been an issue for the researchers (it also happens with France in the case of Europe and some other interesting countries like Myanmar or Eritrea). However Pakistan has never AFAIK cause such kind of red tape trouble, so its undersampling seems quite unjustified.
"In this scenario, how do we expect a fair assessment of R1a1 diversity within South Asia ?"
Your complaint does indeed suggest that there is some potential for further research in the subcontinent.
"... haplogroup M576"...
This marker is not listed by ISOGG. Not sure what it may mean.
AFAIK "M" markers have all been described by Underhill's lab, so I would suggest that you politely write to their corresponding author asking for more info on the matter. I imagine it is some new sublineage but unsure.
"Afghanistan has been considered a part of Central Asia which in my opinion is not correct."
For me it is the triple knot between these three regions: West, Central and South Asia. There's no clear-cut geographic border in any case. Not only in this case anyhow. Even the Caucasus are a poor geographic border when it comes to segregate Europe and West Asia, the genetic divide being actually further north. Or NE India as buffer between South and East Asia, etc.
Not sure how relevant this may be, if at all.
"I can send you a copy if you provide an email."
DeleteI will be very glad if you do so. My email happens to be rathodhj@yahoo.com
"Your complaint does indeed suggest that there is some potential for further research in the subcontinent."
Thank you for acknowledging that there is legitimacy in my concern. I hope that in the near future a much greater representative sampling of South Asia takes place. And I hope the Indian bureaucracy is not a hindrance.
"AFAIK "M" markers have all been described by Underhill's lab, so I would suggest that you politely write to their corresponding author asking for more info on the matter."
I shall do so. This M576 seems to be very spread out, ranging from Iran, UAE, Oman & even Turkey & 1 sample from Crete & Armenia to Afghanistan, Pakistan and all across India as well as among the Roma.
Thanks & Regards.
Paper sent already.
DeleteHi Jaydeep
DeleteWhile the points you make regarding the sampling is relevant and as maju mentioned here the legal restrictions is the reasons for the lack of samples from that . Despite that from the available information south asia couldnt possibly be the source and higher frequencies among some groups is indeed due to founder effects .
The majority of the south asians come under R-Z94 and downstream subclades .mainly belonging L657(which is equivalent to M576) and Z2125 .
The reason why iran is brought in these discussions is because they do have much older r1a subclades which is seemingly absent in south asia .Even then i dont see indian R1a subclades coming from west asia but rather from central asia . Also eurasian steppes is the place of likely origin for Z645 which is ancestral to Z283 european clade and Z93 asian clade
In any event, this paper's conclusions and yours seem very logical. This is exactly the time and place where goats and cows are believed to have been domesticated. I don't think it's a stretch to posit that R1a carrying pastoralists radiated out from the Zagros mountains starting around ~10Ka. Then a second wave begins with the domestication of the horse on the Pontic-Caspian Steppe, spreading to neighbouring pastoralists and beyond.
ReplyDeleteIt rather elegantly marries the Anatolian hypothesis and the Kurgan hypothesis too. Both are correct, just at different time depths.
It almost fits too well.
I'm rather thinking in a still Mesolithic chronology in fact. Whatever the case it's very difficult to explain immigration from West Asia to Eastern Europe, be it in Mesolithic, Neolithic or Chalcolithic times: no archaeological trail seems to fit and also autosomal DNA, when compared with ancient European and Siberian hunter-gatherers, as well as with European early farmers, seems to support the Paleolithic model for the origins of Eastern Europeans, at least in essence.
DeleteThere are many issues yet to be solved.
You could be right, though I think you and I are looking at a similar time frame just from opposite directions. It could still be a goat-mediated change. Hunting herds of animals in grasslands is a very different game from hunting them in forests. If you're hunting gazelle in grasslands you follow the herds. There's no sense in just sitting in one place once the herds have migrated elsewhere. When you're hunting things like deer, there's suddenly a real danger of overhunting, and you need to protect your hunting grounds from other groups. I'm paraphrasing something I read earlier here and doing a bad job of it... I'll try to find the original source later. My point is that when you go from grasslands to forests as was happening at the time in the Zagros mountains, demographic pressures suddenly appear that weren't there before, and those could drive migration. It could also drive domestication. The archaeological record shows just that sort of shift happening with the Zarzian culture.
DeleteI'm not sure how interested you are in the genetics of goats, but you may want to check out these two articles:
http://www.pnas.org/content/105/46/17659.full.pdf+html
http://www.sciencemag.org/content/287/5461/2254.short
The second is about the oldest domestic goat remains, dated from ~10,000 BP. The first shows that the genetic evidence points to a common ancestor for the vast majority of domestic goats in the world somewhere in the same area of Kurdistan in the Zagros mountains.
Correct me if I'm wrong, but pre-pottery nomads don't leave the same sort of archaeological record as more sedentary groups, do they? In terms of autosomal DNA, were they necessarily all that far apart to begin with? Even if they were, would it be the first time there was a big disconnect from Y-DNA and autosomal DNA? IIRC there was a paper that showed the effective founding population size for men in Europe was really low compared to women.
I don't know if you have this problem in your area, but where I am (British Columbia, though the problem is in several western states in the US as well) there's a community of pseudo-Mormon polygamists, and what they do is basically marry the girls young and kick out the "surplus" boys as soon as they can. I would imagine the spread of a polygamous culture like that change an area's Y-DNA quite quickly. If you look up Bountiful BC (that's actually the name of the town) you can find more info.
Rushed response (busy with work) so sorry if it's a bit... scattered. I can try and find better sources if there's an interest.
I am well aware of goats (and probably also sheep) being domesticated first in the Zagros area although I thank you anyhow for the 2008 study particularly because I was not aware of it.
DeleteWhatever the case, the destination geography of R1a migrants is very different from the Zagros area, at the very least in Europe, so I don't see how to make any easy connection.
"... pre-pottery nomads don't leave the same sort of archaeological record as more sedentary groups, do they?"
It should be recognizable anyhow, provided that enough research has been done.
"In terms of autosomal DNA, were they necessarily all that far apart to begin with?"
Have you read the Lazaridis et al. recent study on ancient European DNA?
→ http://forwhattheywereweare.blogspot.com/2013/12/ancient-european-dna-and-some-debatable.html
It's not the only such data, of course: the general outline was known before with less aDNA info.
"I don't know if you have this problem in your area"...
No. Polygamy is historically very rare in Europe, although illegitimate children with servant and slave women were probably more common once. Whatever the case, if we are talking hunter-gatherers or even early farmers, such a "sophisticated" family system is almost automatically discarded: it rather belongs to the Metal Ages and Feudalism.
Hunter-gatherers, unlike Metal Ages' conquerors, would travel with their full families, at least more often than not. We do have examples (Native Americans, Uralic peoples) who show a clear differential heritage on the paternal and maternal/autosomal side but these changes almost certainly happened after long periods of living side by side with other autochtonous peoples: the Y-DNA was largely retained because of patrilocality while the mtDNA (and autosomal DNA) was diluted almost beyond recognition after continuous unequal admixture with their more populous neighbors to the south.
"It should be recognizable anyhow, provided that enough research has been done."
DeleteWell, the whole are hasn't been the safest place to do research in a while. Most of the articles I came across on the Zarzian culture seem to be from the early 1980s or earlier. Without a solid basis for the source it might be harder to find the evidence elsewhere.
I've read Lazaridis et al. I'm just asking if we know for sure that the early R1a groups were closely related to Anatolians and EEF groups. I think I've seen you mention here before that some suggest the Zarzians repopulated the Zagros from an Epigravettian source in Eastern Europe via the Caucusus. They may have been genetically distinct from the groups directly to the West and South of them.
"I think I've seen you mention here before that some suggest the Zarzians repopulated the Zagros from an Epigravettian source in Eastern Europe via the Caucusus".
DeleteI've mentioned that indeed but my sources for that are old (70s-80s) and I have many doubts if such an interpretation stands today.
In any case, it seems that the pre-M417 R1a belongs to of South/West Asia and that the M417 node implies a fast expansion from (almost certainly) that area, so no need for any European precursor role in fact.
"I'm just asking if we know for sure that the early R1a groups were closely related to Anatolians and EEF groups".
We don't know that much. I'm inferring from Eastern and European modern genetics, which is largely dominated by R1a, that they were most likely not too close to EEFs or West Asians.
Sorry I should have been clearer - when I said early R1a, I meant those pre-M417 R1a. I'm wondering out loud if the Zarzians may have been genetically distinct from the groups living immediately to the south and west, with the region of Kurdistan acquiring its current West Asian genetic identity from later population movements.
DeleteKurds do have greater "Northern European" affinities than their neighbours according to that recent Dodecad K12b run posted on Kurdish DNA. ~5-8%. It would be interesting to know if this is recent admixture or from some older population structure.
"I've mentioned that indeed but my sources for that are old (70s-80s) and I have many doubts if such an interpretation stands today. "
It seems like a lot of research in the region stopped with the outbreak of the Turkish-Kurdish conflict and the Iran-Iraq war unfortunately. Some modern DNA tests on some Zarzian remains would be really interesting to see. Pity.
Would you happen to be aware of any good publications on philogeny of the mDNA clades present in both Kurds and Kurgan remains? I'm thinking particularly haplogroups C4, U5, U3 and T* in particular. Are they sister lineages, or are one derived from the other, and with what sort of timeline? Though even with that information it may be hard to distinguish between deep Epipaleolithic lineages and more recently arrived Iranian and Near Eastern lineages in Kurdistan. Hrm.
The best resource I can think of for Kurdish DNA is the "Kurdish DNA" blog of Palisto. I'm quite sure that there is no aDNA data specifically from Kurdistan but there is some from nearby areas of the Syrian Euphrates and in some cases also from nearby parts of Turkey. I'd use Jean Manco's aDNA list for a first reference (but double-check with listed sources when possible because there are instances, at least in Paleolithic Europe, in which she misreports the data to fit her rather peculiar biases). To check the phylogeny, once you know the sequences, the best reference is PhyloTree (the equivalent of ISOGG for mtDNA). If you find something interesting, please let me know.
DeleteWikipedia argues that the Baltic and Slavonic split occurred 1500-1000 BCE, and the coalescent time estimate for M458 in East/Central Europe is between 8384 and 2314 years and for M558 between 9819 and 2710 years. Ukrainians have the oldest coalescent time estimate for Z282, i.e. between 14795 and 4083 years. Thus, Z282 is quite old to be specifically linked with the proto-IE language.
ReplyDeleteAccording to Wikipedia, ”Baltic languages were spoken over a larger area: West to the mouth of the Vistula river in present-day Poland, at least as far East as the Dniepr river in present-day Belarus, perhaps even to Moscow, perhaps as far south as Kiev. Key evidence of Baltic language presence in these regions is found in hydronyms (names of bodies of water) in the regions that are characteristically Baltic. Historical expansion of the usage of Slavic languages in the South and East, and Germanic languages in the West reduced the geographic distribution of Baltic languages to a fraction of the area that they formerly covered”, and ” the range of the Eastern Balts once reached to the Ural mountains”. I find it also highly interesting that Wikipedia argues that ”more recent scholarship has suggested that there was no unified Proto-Baltic stage, but that Proto-Balto-Slavic split directly into three groups: Slavic, East Baltic and West Baltic. Under this view, the Baltic family is paraphyletic, and consists of all Balto-Slavic languages that are not Slavic. This would imply that Proto-Baltic, the last common ancestor of all Baltic languages, would be identical to Proto-Balto-Slavic itself, rather than distinct from it. Finally, there is a minority of scholars who argue that Baltic descended directly from Proto-Indo-European, without an intermediate common Balto-Slavic stage. They argue that the many similarities and shared innovations between Baltic and Slavic are due to several millennia of contact between the groups, rather than shared heritage”.
I find Table S4 very interesting. When you compare the various haplotype frequencies, you see that the frequencies of M558 and M458 are low in Scandinavia and England (0-2.8%). Conversely, the frequency of Scandinavian Z284 in Eastern Europe and Russia is practically zero. It does not seem that the Vikings left much genetic legacy in Eastern Europe. It is a pity that Balts have been omitted from the comparison. Russians, Belorussians, Hmelnitsk Ukrainians, Hungarians and Estonians carry clearly much more M558 than M458, whereas Czechs and Croats carry definitely much more M458 than M558. It seems that M458 and M558 are both typical for Balto-Slavic populations, but they may have arisen already at the stage of the proto-IE language or even before.
I must say that I feel tempted to argue that M458 is more related to the expansion ofSlavic languages and M558 to the expansion of Proto-Balto-Slavic. It would be exciting to know if Xinjiang Tarim Basin R1a belongs to to Z93. (http://upload.wikimedia.org/wikipedia/commons/4/4f/IndoEuropeanTree.svg)
Volgaic groups seem to have a high frequency of M558 and a low frequency of M458, e.g. Maris, Udmurts, Komi-Permyaks and Chuvashes carry 0% of M458, which seems to indicate that their R1a is not coming from the Russians but precedes the Slavic period.
What R1a-Z282 scatter pattern most suggest the most to me is a combo of Dniepr-Don → Pitted Ware, on one side, and, on the other, IE expansion to Central Europe (Sredny-Stog II → Baalberge → Corded Ware) and back to Eastern Europe (Luboń → Globular Amphorae → Corded Ware).
DeleteThis suggests that IE/Kurgan expansion was not a mere linear process in which one founder lineage marks all the expansion but that they, much as Turks later on, incorporated other populations and locally various founder effects took place with various origins (or in some cases not important obvious genetic impact of any kind).
The dates of the process are in any case incompatible with such a recent phenomenon as the Slavic expansion, which only has about 1300 years of age.
"Conversely, the frequency of Scandinavian Z284 in Eastern Europe and Russia is practically zero. It does not seem that the Vikings left much genetic legacy in Eastern Europe".
It's almost exclusively a Norwegian marked and Norwegians didn't leave much of a mark anywhere (except North of Scotland). When we talk of Vikings we talk of basically the Danish and when we talk of Varangians we talk of the Swedes.
Wikipedia argues that the Baltic and Slavonic split occurred 1500-1000 BCE, and the coalescent time estimate for M458 in East/Central Europe is between 8384 and 2314 years and for M558 between 9819 and 2710 years. Ukrainians have the oldest coalescent time estimate for Z282, i.e. between 14795 and 4083 years. Thus, Z282 is quite old to be specifically linked with the proto-IE language.
ReplyDeleteAccording to Wikipedia, ”Baltic languages were spoken over a larger area: West to the mouth of the Vistula river in present-day Poland, at least as far East as the Dniepr river in present-day Belarus, perhaps even to Moscow, perhaps as far south as Kiev. Key evidence of Baltic language presence in these regions is found in hydronyms (names of bodies of water) in the regions that are characteristically Baltic. Historical expansion of the usage of Slavic languages in the South and East, and Germanic languages in the West reduced the geographic distribution of Baltic languages to a fraction of the area that they formerly covered”, and ” the range of the Eastern Balts once reached to the Ural mountains”. I find it also highly interesting that Wikipedia argues that ”more recent scholarship has suggested that there was no unified Proto-Baltic stage, but that Proto-Balto-Slavic split directly into three groups: Slavic, East Baltic and West Baltic. Under this view, the Baltic family is paraphyletic, and consists of all Balto-Slavic languages that are not Slavic. This would imply that Proto-Baltic, the last common ancestor of all Baltic languages, would be identical to Proto-Balto-Slavic itself, rather than distinct from it. Finally, there is a minority of scholars who argue that Baltic descended directly from Proto-Indo-European, without an intermediate common Balto-Slavic stage. They argue that the many similarities and shared innovations between Baltic and Slavic are due to several millennia of contact between the groups, rather than shared heritage”.
I find Table S4 very interesting. When you compare the various haplotype frequencies, you see that the frequencies of M558 and M458 are low in Scandinavia and England (0-2.8%). Conversely, the frequency of Scandinavian Z284 in Eastern Europe and Russia is practically zero. It does not seem that the Vikings left much genetic legacy in Eastern Europe. It is a pity that Balts have been omitted from the comparison. Russians, Belorussians, Hmelnitsk Ukrainians, Hungarians and Estonians carry clearly much more M558 than M458, whereas Czechs and Croats carry definitely much more M458 than M558. It seems that M458 and M558 are both typical for Balto-Slavic populations, but they may have arisen already at the stage of the proto-IE language or even before.
I must say that I feel tempted to argue that M458 is more related to the expansion ofSlavic languages and M558 to the expansion of Proto-Balto-Slavic. It would be exciting to know if Xinjiang Tarim Basin R1a belongs to to Z93. (http://upload.wikimedia.org/wikipedia/commons/4/4f/IndoEuropeanTree.svg)
Volgaic groups seem to have a high frequency of M558 and a low frequency of M458, e.g. Maris, Udmurts, Komi-Permyaks and Chuvashes carry 0% of M458, which seems to indicate that their R1a is not coming from the Russians but precedes the Slavic period.
True, but there is Z284 in North England, Sweden and Denmark: North England 3.4%, Denmark 7.1%, South Sweden 3.5%. In Germany there seems to be only 0.9%.
ReplyDeleteThe oldest time estimate for M458 is in Poles and the oldest time estimate for M558 is in Slovaks, which means that the areas are geographically very close. Z284 seems to be older in Ukraina than in Russia or Belorussia. If the Dniepr-Don culture radiated northward during the Neolithic, it was probably not an IE culture in the strict sense of the word. I do not believe that the Pitted Ware (ca 3200 BC– ca 2300 BC) people spoke an IE language, unless we think that the IE languages are a wide areal phenomenon with various substrates and which developed as a result of the post-Ice Age expansion of people from Eastern Europe. If the origin of IE languages is in Sredny Stog culture (4000-3500 BC), we should postulate a migration of M458 and M558 from Ukraine to Czech and Poland in the form of the Corded Ware. However, it is possible that Z284, M458 and M558 were in Central Europe already before the Corded Ware and preceded any proper IE language. However, as they all may have their origin in the area of Ukraine, it is possible that these languages were similar to the proper IE languages and were some sort of para-IE languages.
Wikipedia article on Pitted Ware even speculates that "as the (Pitted Ware) language left no records, its linguistic affiliations are uncertain. It has been suggested that its people spoke a language related to the Uralic languages and provided the unique linguistic features discussed in the Germanic substrate hypothesis."
Anyway, I think that the IE languages arrived to Scandinavia with the Corded Ware. According to Wikipedia, the Corded Ware culture flourished in Middle Europe c. 2900 – 2450/2350 cal. BC. Around 2400 BC the people of the Corded Ware replaced their predecessors and expanded to Danubian and Nordic areas of western Germany. A related branch invaded Denmark and southern Sweden.
I do not believe in this replacement of yDNA, but think that at this stage the IE yDNA had already merged with the local yDNA in the area of Germany and Poland. Moreover, IMO, part of Z282 had spread to Northern Europe before the Corded Ware and may have spoken whatever European paleo-languages. As Y DNA wise, Germanic and Slavic areas are quite distinct, there can hardly be said to have occurred any Bronze Age Indo-European yDNA replacement, in particular, in Scandinavia.
"there is Z284 in North England, Sweden and Denmark: North England 3.4%, Denmark 7.1%, South Sweden 3.5%. In Germany there seems to be only 0.9%".
ReplyDeleteThat's interesting. I guess that if you'd take as "Viking" marker (or Viking+Anglosaxon), it would imply that almost half of North English patrilineal ancestry is from that area (based on the Danish frequency). But it's also possible that it is something quite older in the region, maybe from the time of Hamburgian or Maglemosian cultures, which spanned across the North Sea.
"The oldest time estimate for M458 is in Poles and the oldest time estimate for M558 is in Slovaks, which means that the areas are geographically very close".
I wouldn't trust so much localized age estimates without considering the phylogeny (it can be an artifact of immigration from various sources). If we would know that the basal nodes and branches of these subhaplogroups are concentrated in those areas, I would accept the origin hypothesis but the haplotype network is not so precise about location, so it needs of much extra work in order to find out the exact geography of the expansion of these branches. The data is there anyhow, so I guess it's just a matter of building the haplotype network with the necessary country labels instead of just "Eastern European" for all.
"If the Dniepr-Don culture radiated northward during the Neolithic, it was probably not an IE culture in the strict sense of the word".
Surely not. But its radiation may have set some of the genetic basis on which the IE wave rode later on. After all DD was the first "victim" of Kurgan expansion but in a very complex way: Sredny-Stog II was a patchy society that mixes both traditions in very irregular forms. Some cultures that the Kurgan expansion produced, like Ezero (proto-Thracians), actually retained many DD cultural traits (extended burial with ochre and such), while in the Baltic Pitted Ware was in a way opening the route of later Kurgan flows. So it does not surprise me that DD peoples may have manned the Kurgan expansion even if they were originally distinct from IEs (how distinct?)
"I do not believe that the Pitted Ware (ca 3200 BC– ca 2300 BC) people spoke an IE language"...
Maybe they spoke a language distantly related to PIE. We do not know anything about the Neolithic genesis of the Samara culture but it seems apparent that they had at least some influence from DD.
IF the hypothesis that Vasconic is distantly related to PIE is correct, and my theory of Vasconic being spread by Neolithic Farmers stands, then it is indeed possible that there was once a linguistic family in SE-Eastern Europe that gave birth to Vasconic, PIE and surely other branches now lost, very possibly the language of Dniepr-Don and Pitted Ware peoples. Not demonstrated but quite plausible.
But I agree that it's not likely that PIE included the DD language as such. However it is very possible that the DD language strongly influenced the Western branches of IE as substrate initially, after all they used the Dniepr-Don area as main platform for further intrusions westwards.
...
...
Delete"we should postulate a migration of M458 and M558 from Ukraine to Czech and Poland in the form of the Corded Ware".
Don't forget the precursors: Baalberge →→ Luboń→ Globular Amphorae → Corded Ware. It is true that at the genesis of CW there is an intrusive element from Catacombs culture but otherwise the localized Central-East European genesis pattern stands. So, yes, there is a Sredny-Stog II →→→ Corded Ware link without doubt but mediated by almost a millennium of local North-Central European development between East Germany and NW Ukraine, largely centered in Poland.
" It has been suggested that its people spoke a language related to the Uralic languages and provided the unique linguistic features discussed in the Germanic substrate hypothesis".
I'd rather suspect their influence to be found in the Baltic languages but whatever.
"IE languages arrived to Scandinavia with the Corded Ware."
Yes indeed. That was the moment of IE consolidation east of the Rhine.
"at this stage the IE yDNA had already merged with the local yDNA in the area of Germany and Poland."
Exactly: a whole millennium of consolidation would have converted all the local substrate (Danubian, Funnelbeaker, even the last remnant hunter-gatherers) into Indoeuropeans.
"there can hardly be said to have occurred any Bronze Age Indo-European yDNA replacement, in particular, in Scandinavia".
If you look at the archaeo-demographic evidence, it seems quite apparent that, barring some very localized exceptions, there was no demographic growth at the arrival of CW. That helps explain their limited genetic impact. On the other hand the previous Megalithic phenomenon marked major expansions and was surely therefore leaving a more important genetic legacy.
BTW, Ethio Helix just posted a new entry in which he creates haplotype networks using the Network software (free, CC license).
DeleteSomeone really interested in the Z282 exact origins could probably use it to produce informative graphs.
@Maju
ReplyDeleteSo where did R1a and R1b split.?..........same place.
According to David Adams, from the blacksea north caucasus through the caspian sea and through to the Aral sea was still underwater due to the melting of the northern central asian . According to Greek historians, the Greeks could still sail to the Aral sea in the bronze-age. With this in mind, your scenario would mean most groups that came from HIJKLT would fit this Iranian scenario
"So where did R1a and R1b split.?"
DeleteSomewhere in West Asia or maybe Pakistan (where most R1* is).
"...same place".
Roughly so. It would seem that Q1, R1b and R1a are just different expansive waves of the same meta-population, each one characterized by a different founder effect.
"... the Greeks could still sail to the Aral sea in the bronze-age".
Can't they now? They can indeed, just as they did back then.
"With this in mind, your scenario would mean most groups that came from HIJKLT would fit this Iranian scenario"
Do you mean F? F(xG)? Not sure what you have in mind. Personally I understand that R1, Q, G, IJ and T migrated westwards (possibly together with C6 and some F*) in the very first Upper Paleolithic. Afterwards they and their descendants had different fortunes but most expanded somewhere at one moment or another (otherwise they would probably have no recognizable name today).
It's of course possible that (pre-)G and/or IJ may have migrated westwards before but we can't really say much about that, only that their ancestor F looks as expanding from South Asia, so they must have migrated westwards at some point anyhow.
Well at least this will end the nonsense of some in South Asia who are advocating a Out of India Theory (OIT) for R1a1, now we know it may be Iran or someplace in West Asia that's the origin.
ReplyDeleteso r1a farmers from middle east bring the wheat farming in india?
ReplyDeleteIn a very oversimplified way, yes.
DeleteI'd say more nuancedly (see: http://forwhattheywereweare.blogspot.com/2014/05/south-asian-first-neolithic-and-its.html) that farming and herding spread from the Fertile Crescent to Iran (then better identified as Elam), from there to Balochistan and Central Asia, then to Pakistan and NW India (eventually leading to IVC) and then, especially after the incorporation of tropical crops, to further India.
The Western neolithic package is much more than just wheat (other cereals, various types of legumes, four types of livestock) and also the patrilineages carried were more than just R1a, at the very least also J2, and from Pakistan to the East it is important to underline L as well. These were also carried to Central Asia, which had a less famous but similarly old Neolithic.
Here is a simple test. How old is the oldest r1a? How old is the oldest r1a found in europe? How old is the oldest r1a found in central asia? How old is the oldest r1a in india? How old is the oldest r1a found in iran?
ReplyDeleteWe don't know for sure: the academic "molecular clock" has been broken for more than a decade, as they keep calibrations made with very obsolete 20th century archaeology such as imagining the OoA c. 60 Ka BP instead of more than 100 Ka BP, or (paleontology rather than archaeology but same thing overall) imagining the Pan-Homo split c. 5-6 Ma ago instead of the at least 8 Ma ago, maybe as much as 17 Ma ago.
DeleteSo we don't know that: the "molecular clock" is infinitely more unreliable than C14.
Also, in order to find, you have to search. Europe is over-studied, Asia, Africa, etc. are not.
Delete